Uncovering latent infections in kidneys: A novel molecular approach for differential Leptospira detection
- PMID: 39691702
- PMCID: PMC11650133
- DOI: 10.1016/j.crmicr.2024.100327
Uncovering latent infections in kidneys: A novel molecular approach for differential Leptospira detection
Abstract
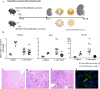
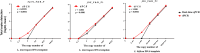
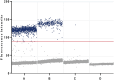
Leptospirosis, a re-emerging zoonotic disease caused by Leptospira spp., poses significant global health and veterinary challenges. Long-term colonization of renal tubules by Leptospira in asymptomatic hosts highlights the need for sensitive detection methods. This study evaluates the chronic or latent Leptospira infections in kidneys using a novel molecular approach to examine individual immune responses differences. Digital PCR strategies employing newly developed primer-probe sets targeting the flagellar fliG gene were used to assess the presence of trace Leptospira in infected murine kidneys and urine samples from laboratory-confirmed leptospirosis patients. RNA-based digital PCR detected leptospires in 58 % (targeting lipl32) and 83 % (targeting fliG) of infected kidneys, demonstrating that the digital PCR strategy targeting the fliG gene offers superior sensitivity. Notably, the newly developed fliG-targeting assay detected as low as 20 fg of Leptospira DNA, offering ten-fold greater sensitivity than traditional qPCR for trace detection. This allows for differential detection of Leptospira species and facilitates monitoring of extremely low bacterial loads with greater sensitivity than conventional methods. We also observed regenerating renal tubules with mitosis and elevated cytokine expression in kidneys with transcriptionally active Leptospira during chronic infection. This approach aids in identifying latent infections and offers insights into individual variations. Our research provides a powerful molecular tool for epidemiological studies and public health surveillance, contributing valuable insights into the prevalence and transmission dynamics of this pervasive zoonotic disease.
Keywords: Differential detection; Digital PCR; Latent infection; Leptospira spp.; Leptospirosis; Leptospirosis kidney disease.
© 2024 The Author(s).
Conflict of interest statement
L.-F. C., P.-Y. H. and C.-W. Y. are inventors (Kidney Research Center, Chang Gung Memorial Hospital) on a patent for “Innovative method for nucleic acid detection tracking minute quantities of Leptospira spp. in biological tissues and environmental samples” (US Provisional Patent Application 63/605,750, filed 4 December 2023). The remaining authors declare no competing interests.
Figures




References
-
- Bárbara Couto Roloff Padilha M.M.S., Daiane Drawanz Hartwig Molecular and serological diagnostic of leptospirosis: a review (2014–2020) Res, Soc Dev. 2022;11 doi: 10.33448/rsd-v11i2.25471. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

