A blood-based mRNA signature distinguishes people with Long COVID from recovered individuals
- PMID: 39691709
- PMCID: PMC11649547
- DOI: 10.3389/fimmu.2024.1450853
A blood-based mRNA signature distinguishes people with Long COVID from recovered individuals
Abstract
Introduction: Long COVID is a debilitating condition that lasts for more than three months post-infection by SARS-CoV-2. On average, one in ten individuals infected with SARS CoV- 2 develops Long COVID worldwide. A knowledge gap exists in our understanding of the mechanisms, genetic risk factors, and biomarkers that could be associated with Long COVID.
Methods: In this pilot study we used RNA-Seq to quantify the transcriptomes of peripheral blood mononuclear cells isolated from COVID-recovered individuals, seven with and seven without Long COVID symptoms (age- and sex-matched individuals), on average 6 months after infection.
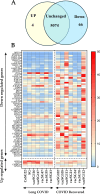
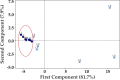
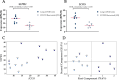
Results: Seventy genes were identified as significantly up- or down-regulated in Long COVID samples, and the vast majority were downregulated. The most significantly up- or downregulated genes fell into two main categories, either associated with cell survival or with inflammation. This included genes such as ICOS (FDR p = 0.024) and S1PR1 (FDR p = 0.019) that were both up-regulated, indicating that a pro-inflammatory state is sustained in Long COVID PBMCs compared with COVID recovered PBMCs. Functional enrichment analysis identified that immune-related functions were expectedly predominant among the up- or down-regulated genes. The most frequently downregulated genes in significantly altered functional categories were two leukocyte immunoglobulin like receptors LILRB1 (FDR p = 0.005) and LILRB2 (FDR p = 0.027). PCA analysis demonstrated that LILRB1 and LILRB2 expression discriminated all of the Long COVID samples from COVID recovered samples.
Discussion: Downregulation of these inhibitory receptors similarly indicates a sustained pro-inflammatory state in Long COVID PBMCs. LILRB1 and LILRB2 should be validated as prospective biomarkers of Long COVID in larger cohorts, over time and against clinically overlapping conditions.
Keywords: COVID-19; LILRB1; LILRB2; Long COVID; biomarker; inflammation; transcriptomics.
Copyright © 2024 Missailidis, Ebrahimie, Dehcheshmeh, Allan, Sanislav, Fisher, Gras and Annesley.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

