Expansion of the MutS gene family in plants
- PMID: 39692564
- PMCID: PMC12292046
- DOI: 10.1093/plcell/koae277
Expansion of the MutS gene family in plants
Abstract
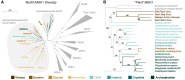
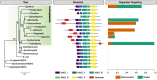
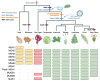
The widely distributed MutS gene family functions in recombination, DNA repair, and protein translation. Multiple evolutionary processes have expanded this gene family in plants relative to other eukaryotes. Here, we investigate the origins and functions of these plant-specific genes. Cyanobacterial-like MutS1 and MutS2 genes were ancestrally gained via plastid endosymbiotic gene transfer. MutS1 was subsequently lost in seed plants, whereas MutS2 was duplicated in Viridiplantae (i.e. land plants and green algae). Viridiplantae also have 2 anciently duplicated copies of the eukaryotic MSH6 gene and acquired MSH1 via horizontal gene transfer-potentially from a nucleocytovirus. Despite sharing a name, "plant MSH1" is not directly related to the MSH1 gene in some fungi and animals, which may be an ancestral eukaryotic gene acquired via mitochondrial endosymbiosis and subsequently lost in most eukaryotes. There has been substantial progress in understanding the functions of plant MSH1 and MSH6 genes, but the cyanobacterial-like MutS1 and MutS2 genes remain uncharacterized. Known functions of bacterial homologs and predicted protein structures, including fusions to diverse nuclease domains, provide hypotheses about potential molecular mechanisms. Because most plant-specific MutS proteins are mitochondrial and/or plastid-targeted, the expansion of this family has played a large role in shaping plant organelle genetics.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures



Update of
-
Expansion of the MutS Gene Family in Plants.bioRxiv [Preprint]. 2024 Jul 20:2024.07.17.603841. doi: 10.1101/2024.07.17.603841. bioRxiv. 2024. Update in: Plant Cell. 2025 Jul 1;37(7):koae277. doi: 10.1093/plcell/koae277. PMID: 39071318 Free PMC article. Updated. Preprint.
References
-
- Abdelnoor R. Cloning and characterization of MSH1 in higher plants and its involvement in regulation of substoichiometric shifting. Lincoln, NE: University of Nebraska; 2004.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

