Deep learning detected histological differences between invasive and non-invasive areas of early esophageal cancer
- PMID: 39692707
- PMCID: PMC11875758
- DOI: 10.1111/cas.16426
Deep learning detected histological differences between invasive and non-invasive areas of early esophageal cancer
Abstract
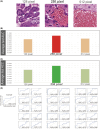
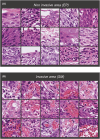
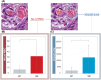
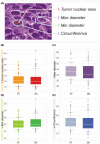
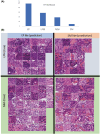
The depth of invasion plays a critical role in predicting the prognosis of early esophageal cancer, but the reasons behind invasion and the changes occurring in invasive areas are still not well understood. This study aimed to explore the morphological differences between invasive and non-invasive areas in early esophageal cancer specimens that have undergone endoscopic submucosal dissection (ESD), using artificial intelligence (AI) to shed light on the underlying mechanisms. In this study, data from 75 patients with esophageal squamous cell carcinoma (ESCC) were analyzed and endoscopic assessments were conducted to determine submucosal (SM) invasion. An AI model, specifically a Clustering-constrained Attention Multiple Instance Learning model (CLAM), was developed to predict the depth of cancer by training on surface histological images taken from both invasive and non-invasive regions. The AI model highlighted specific image portions, or patches, which were further examined to identify morphological differences between the two types of areas. The 256-pixel AI model demonstrated an average area under the receiver operating characteristic curve (AUC) value of 0.869 and an accuracy (ACC) of 0.788. The analysis of the AI-identified patches revealed that regions with invasion (SM) exhibited greater vascularity compared with non-invasive regions (epithelial). The invasive patches were characterized by a significant increase in the number and size of blood vessels, as well as a higher count of red blood cells (all with p-values <0.001). In conclusion, this study demonstrated that AI could identify critical differences in surface histopathology between non-invasive and invasive regions, particularly highlighting a higher number and larger size of blood vessels in invasive areas.
Keywords: artificial intelligence; epithelium; esophageal cancer; submucosa; surface histomorphology.
© 2024 The Author(s). Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
Dr. Genichiro Ishii is an Editorial Board member of
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

