Comparison of targeted next generation sequencing assays in non-small cell lung cancer patients
- PMID: 39692940
- PMCID: PMC11655926
- DOI: 10.1007/s12672-024-01640-7
Comparison of targeted next generation sequencing assays in non-small cell lung cancer patients
Abstract
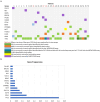
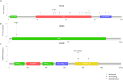
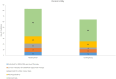
Non-small cell lung cancer (NSCLC) is the most prevalent type of lung cancer the mutational spectrum of which has been extensively characterized. Treatment of patients with NSCLC based on their molecular profile is now part of the standard clinical care. The aim of this study was firstly to investigate two different NGS-based tumor profile genetic tests and secondly to assess the clinical actionability of the mutations and their association with survival and clinicopathological characteristics. Overall, 52 mutations were identified in 31 patients by either one or both assays. The most frequently mutated genes were TP53 (40.4%), KRAS (13.46%) and EGFR (9.62%). TP53 and KRAS mutations were associated with worst overall survival while KRAS was positively correlated with adenocarcinoma. The two methods showed a high concordance for the commonly covered genomic regions (97.14%). Ten mutations were identified in a genomic region exclusively covered by the MEDICOVER Genetics custom tumor profile assay. Likewise, one MET mutation was identified by the Ion Amliseq assay in a genomic region exclusively covered by Ion Amliseq. In conclusion both assays showed highly similar results in the commonly covered genomic areas, however, the MEDICOVER Genetics assay identified additional clinically actionable mutations that can be applied in clinical practice for personalized treatment decision making for patients with NSCLC.
Keywords: clinical utility; mutational profile; next generation sequencing; NSCLC.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: CL: Employed by MEDICOVER Genetics has filed a PCT patent application for Target-enriched multiplexed parallel analysis for assessment of tumor biomarkers (WO2019/008172A1); AA: Employed by MEDICOVER Genetics; has filed a PCT patent application for the Target-enriched multiplexed parallel analysis for assessment of tumor biomarkers (WO2019/008172A1); has filed a PCT patent application for the Enrichment of Targeted Genomic Regions for Multiplexed Par-allel Analysis (WO2019/008148A9); EK: Employed by MEDICOVER Genetics; has filed a PCT pa-tent application for the Target-enriched multiplexed parallel analysis for assessment of tumor biomarkers (WO2019/008172A1); has filed a PCT patent application for the Enrichment of Tar-geted Genomic Regions for Multiplexed Parallel Analysis (WO2019/008148A9); KT: Employed by MEDICOVER Genetics, has filed a PCT patent application for the Enrichment of Targeted Ge-nomic Regions for Multiplexed Parallel Analysis (WO2019/008148A9); AE: Employed by MEDI-COVER Genetics; has filed a PCT patent application for the Target-enriched multiplexed parallel analysis for assessment of tumor biomarkers (WO2019/008172A1); MI: Employed by MEDICOV-ER Genetics; has filed a PCT patent application for the Target-enriched multiplexed parallel analysis for assessment of tumor biomarkers (WO2019/008172A1), has filed a PCT patent applica-tion for the Enrichment of Targeted Genomic Regions for Multiplexed Parallel Analysis (WO2019/008148A9); GK: Employed by MEDICOVER Genetics; has filed a PCT patent application for the Target-enriched multiplexed parallel analysis for assessment of tumor biomarkers (WO2019/008172A1); has filed a PCT patent application for the Enrichment of Targeted Genomic Regions for Multiplexed Parallel Analysis (WO2019/008148A9); PCP: Employed by MEDICOVER Genetics; has filed a PCT patent application for the Target-enriched multiplexed parallel analysis for assessment of tumor biomarkers (WO2019/008172A1); has filed a PCT patent application for the Enrichment of Targeted Genomic Regions for Multiplexed Parallel Analysis (WO2019/008148A9); The rest of the authors declare no conflict of interest.
Figures



References
-
- Bray F, et al. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. 10.3322/caac.21492. - PubMed
-
- De Perrot M, et al. Sex differences in presentation, management, and prognosis of patients with non-small cell lung carcinoma. J Thorac Cardiovasc Surg. 2000. 10.1016/S0022-5223(00)70213-3. - PubMed
-
- Ettinger DS, et al. NCCN guidelines insights: non-small cell lung cancer, version 1.2020. J Natl Compr Cancer Netw. 2019. 10.6004/jnccn.2019.0059. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
