Detecting gene expression in Caenorhabditis elegans
- PMID: 39693264
- PMCID: PMC11979774
- DOI: 10.1093/genetics/iyae167
Detecting gene expression in Caenorhabditis elegans
Abstract
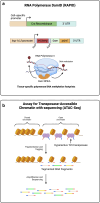
Reliable methods for detecting and analyzing gene expression are necessary tools for understanding development and investigating biological responses to genetic and environmental perturbation. With its fully sequenced genome, invariant cell lineage, transparent body, wiring diagram, detailed anatomy, and wide array of genetic tools, Caenorhabditis elegans is an exceptionally useful model organism for linking gene expression to cellular phenotypes. The development of new techniques in recent years has greatly expanded our ability to detect gene expression at high resolution. Here, we provide an overview of gene expression methods for C. elegans, including techniques for detecting transcripts and proteins in situ, bulk RNA sequencing of whole worms and specific tissues and cells, single-cell RNA sequencing, and high-throughput proteomics. We discuss important considerations for choosing among these techniques and provide an overview of publicly available online resources for gene expression data.
Keywords: FISH; RNA sequencing; WormBook; gene expression; live-cell reporters; single-cell RNA sequencing.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The authors declare no competing interests.
Figures
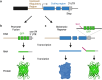

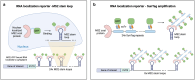


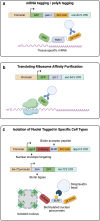

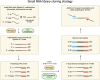






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

