Deep representation learning of protein-protein interaction networks for enhanced pattern discovery
- PMID: 39693438
- PMCID: PMC11654695
- DOI: 10.1126/sciadv.adq4324
Deep representation learning of protein-protein interaction networks for enhanced pattern discovery
Abstract
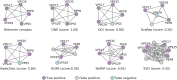
Protein-protein interaction (PPI) networks, where nodes represent proteins and edges depict myriad interactions among them, are fundamental to understanding the dynamics within biological systems. Despite their pivotal role in modern biology, reliably discerning patterns from these intertwined networks remains a substantial challenge. The essence of the challenge lies in holistically characterizing the relationships of each node with others in the network and effectively using this information for accurate pattern discovery. In this work, we introduce a self-supervised network embedding framework termed discriminative network embedding (DNE). Unlike conventional methods that primarily focus on direct or limited-order node proximity, DNE characterizes a node both locally and globally by harnessing the contrast between representations from neighboring and distant nodes. Our experimental results demonstrate DNE's superior performance over existing techniques across various critical network analyses, including PPI inference and the identification of protein functional modules. DNE emerges as a robust strategy for node representation in PPI networks, offering promising avenues for diverse biomedical applications.
Figures




References
-
- Barabasi A.-L., Oltvai Z. N., Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 5, 101–113 (2004). - PubMed
-
- Alon U., Biological networks: The tinkerer as an engineer. Science 301, 1866–1867 (2003). - PubMed
-
- Camacho D. M., Collins K. M., Powers R. K., Costello J. C., Collins J. J., Next-generation machine learning for biological networks. Cell 173, 1581–1592 (2018). - PubMed
-
- Bonneau R., Learning biological networks: From modules to dynamics. Nat. Chem. Biol. 4, 658–664 (2008). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

