A global comparison of surface and subsurface microbiomes reveals large-scale biodiversity gradients, and a marine-terrestrial divide
- PMID: 39693444
- PMCID: PMC11654699
- DOI: 10.1126/sciadv.adq0645
A global comparison of surface and subsurface microbiomes reveals large-scale biodiversity gradients, and a marine-terrestrial divide
Abstract
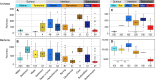
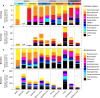
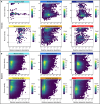
Subsurface environments are among Earth's largest habitats for microbial life. Yet, until recently, we lacked adequate data to accurately differentiate between globally distributed marine and terrestrial surface and subsurface microbiomes. Here, we analyzed 478 archaeal and 964 bacterial metabarcoding datasets and 147 metagenomes from diverse and widely distributed environments. Microbial diversity is similar in marine and terrestrial microbiomes at local to global scales. However, community composition greatly differs between sea and land, corroborating a phylogenetic divide that mirrors patterns in plant and animal diversity. In contrast, community composition overlaps between surface to subsurface environments supporting a diversity continuum rather than a discrete subsurface biosphere. Differences in microbial life thus seem greater between land and sea than between surface and subsurface. Diversity of terrestrial microbiomes decreases with depth, while marine subsurface diversity and phylogenetic distance to cultured isolates rivals or exceeds that of surface environments. We identify distinct microbial community compositions but similar microbial diversity for Earth's subsurface and surface environments.
Figures





References
-
- Takai K., Nakamura K., Toki T., Tsunogai U., Miyazaki M., Miyazaki J., Hirayama H., Nakagawa S., Nunoura T., Horikoshi K., Cell proliferation at 122°C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc. Natl. Acad. Sci. U.S.A. 105, 10949–10954 (2008). - PMC - PubMed
-
- Edgcomb V. P., Molyneaux S. J., Böer S., Wirsen C. O., Saito M., Atkins M. S., Lloyd K., Teske A., Survival and growth of two heterotrophic hydrothermal vent archaea, Pyrococcus strain GB-D and Thermococcus fumicolans, under low pH and high sulfide concentrations in combination with high temperature and pressure regimes. Extremophiles. 11, 329–342 (2007). - PubMed
-
- Ando N., Barquera B., Bartlett D. H., Boyd E., Burnim A. A., Byer A. S., Colman D., Gillilan R. E., Gruebele M., Makhatadze G., Royer C. A., Shock E., Wand A. J., Watkins M. B., The molecular basis for life in extreme environments. Annu. Rev. Biophys. 50, 343–372 (2021). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

