Stratified Medicine Pediatrics: Cell-Free DNA and Serial Tumor Sequencing Identifies Subtype-Specific Cancer Evolution and Epigenetic States
- PMID: 39693475
- PMCID: PMC11962403
- DOI: 10.1158/2159-8290.CD-24-0916
Stratified Medicine Pediatrics: Cell-Free DNA and Serial Tumor Sequencing Identifies Subtype-Specific Cancer Evolution and Epigenetic States
Abstract
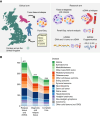
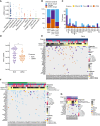
In tumors of childhood, we identify mutations in epigenetic genes as drivers of relapse, with matched cfDNA sequencing showing significant intratumor genetic heterogeneity and cell-state specific patterns of chromatin accessibility. This highlights the power of cfDNA analysis to identify both genetic and epigenetic drivers of aggressive disease in pediatric cancers.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
S.L. George reports other support from Recordati outside the submitted work. C.M. Sauer reports grants and personal fees from Marie Skłodowska-Curie Actions during the conduct of the study. M. Oostveen reports grants from Cancer Research UK during the conduct of the study. S.C. Clifford reports grants from Cancer Research UK during the conduct of the study. D.A. Tweddle reports grants from Cancer Research UK/Blood Cancer UK during the conduct of the study. G.A.A. Burke reports other support from Novartis and AstraZeneca outside the submitted work. J. Turnbull reports other support from government during the conduct of the study. S. Wilson reports personal fees from Norgine and Bayer outside the submitted work. S.A. Gatz reports other support from EMD Serono/Merck KgaA, Amgen, Gilead, GSK, and Schroedinger Therapeutics, grants and other support from AstraZeneca, and grants from Bayer outside the submitted work. L.V. Marshall reports grants from Cancer Research UK during the conduct of the study. T.A. Graham reports other support from DAiNA Therapeutics and Genentech outside the submitted work, as well as a patent for GB2305655.9 pending and a patent for GB2317139.0 pending. B. Al-Lazikani reports grants from Cancer Research UK during the conduct of the study, as well as other support from Excientia PLC now part of Recursion Pharmaceuticals, AstraZeneca, GSK, Astex pharmaceuticals, Stante Ventures, and Astellas Pharmaceuticals outside the submitted work; serving as a member of the scientific committee for Fight Kids Cancer. P. Kearns reports grants from Cancer Research UK during the conduct of the study. D. Hargrave reports personal fees from Novartis, Day One Therapeutics, Ipsen, Biodexa, and Kestrel Therapeuatics and grants and personal fees from Alexion/AstraZeneca outside the submitted work. T.S. Jacques reports grants from Cancer Research UK during the conduct of the study, as well as The Brain Tumor Charity, NIHR, The Olivia Hodson Cancer Foundation, and Children with Cancer UK, other support from Neuropath Ltd and Repath Ltd, and personal fees from Wiley and Elsevier outside the submitted work. M. Hubank reports personal fees from AstraZeneca, Janssen, Alira Health, Servier, Seagen, Qiagen, Roche, and Novartis and nonfinancial support from Guardant Health outside the submitted work. L. Chesler reports grants from Cancer Research UK, Children with Cancer UK, Christopher’s Smile, Abbie’s Fund, and Aoife’s Bubbles and other support from HEFCE/RAE during the conduct of the study. No disclosures were reported by the other authors.
Figures




References
-
- Ceschel S, Casotto V, Valsecchi MG, Tamaro P, Jankovic M, Hanau G, et al. . Survival after relapse in children with solid tumors: a follow-up study from the Italian off-therapy registry. Pediatr Blood Cancer 2006;47:560–6. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

