Near telomere-to-telomere genome assembly of Mongolian cattle: implications for population genetic variation and beef quality
- PMID: 39693631
- PMCID: PMC11653892
- DOI: 10.1093/gigascience/giae099
Near telomere-to-telomere genome assembly of Mongolian cattle: implications for population genetic variation and beef quality
Abstract
Background: Mongolian cattle, a unique breed indigenous to China, represent valuable genetic resources and serve as important sources of meat and milk. However, there is a lack of high-quality genomes in cattle, which limits biological research and breeding improvement.
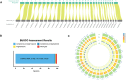
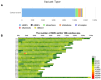
Findings: In this study, we conducted whole-genome sequencing on a Mongolian bull. This effort yielded a 3.1 Gb Mongolian cattle genome sequence, with a BUSCO integrity assessment of 95.9%. The assembly achieved both contig N50 and scaffold N50 values of 110.9 Mb, with only 3 gaps identified across the entire genome. Additionally, we successfully assembled the Y chromosome among the 31 chromosomes. Notably, 3 chromosomes were identified as having telomeres at both ends. The annotation data include 54.31% repetitive sequences and 29,794 coding genes. Furthermore, a population genetic variation analysis was conducted on 332 individuals from 56 breeds, through which we identified variant loci and potentially discovered genes associated with the formation of marbling patterns in beef, predominantly located on chromosome 12.
Conclusions: This study produced a genome with high continuity, completeness, and accuracy, marking the first assembly and annotation of a near telomere-to-telomere genome in cattle. Based on this, we generated a variant database comprising 332 individuals. The assembly of the genome and the analysis of population variants provide significant insights into cattle evolution and enhance our understanding of breeding selection.
Keywords: Mongolian cattle; beef quality; near telomere-to-telomere genome; population genetic variation.
© The Author(s) 2024. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Fedotova GV, Slozhenkina MI, Tsitsige, et al. Comparative analysis of economic and biological features of Kalmyk and Mongolian cattle breeds. IOP Conference Series Earth Environ Sci. 2020;548:082076. 10.1088/1755-1315/548/8/082076. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

