LKB1 dictates sensitivity to immunotherapy through Skp2-mediated ubiquitination of PD-L1 protein in non-small cell lung cancer
- PMID: 39694700
- PMCID: PMC11660338
- DOI: 10.1136/jitc-2024-009444
LKB1 dictates sensitivity to immunotherapy through Skp2-mediated ubiquitination of PD-L1 protein in non-small cell lung cancer
Abstract
Background: Loss-of-function mutations of liver kinase B (LKB1, also termed as STK11 (serine/threonine kinase 11)) are frequently detected in patients with non-small cell lung cancer (NSCLC). The LKB1 mutant NSCLC was refractory to almost all the antitumor treatments, including programmed cell death protein 1 (PD-1)/programmed death-ligand 1 (PD-L1) blockade therapy. Unfortunately, mechanisms underlying resistance to immunotherapy are not fully understood. In this study, we deciphered how LKB1 regulated sensitivity to anti-PD-1/PD-L1 immunotherapy.
Methods: We investigated the mutational landscape of LKB1 mutant NSCLC in next generation sequencing (NGS) data sets. Expression of LKB1, PD-L1 and S-phase kinase-associated protein 2 (Skp2) in NSCLC samples were assessed by immunohistochemistry (IHC). The tumor microenvironment (TME) profiling of LKB1 wild type (WT) and mutant NSCLC was performed using fluorescent multiplex IHC. Mass spectrometry and enrichment analysis were used to identify LKB1 interacting proteins. Mechanistic pathways were explored by immunoblotting, ubiquitination assay, cycloheximide chase assay and immunoprecipitation assay.
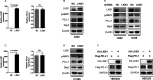
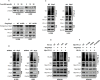
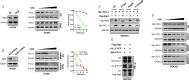
Results: By using NGS data sets and histological approaches, we demonstrated that LKB1 status was positively associated with PD-L1 protein expression and conferred a T cell-enriched "hot" TME in NSCLC. Patients with good responses to anti-PD-1/PD-L1 immunotherapy possessed a high level of LKB1 and PD-L1. Skp2 emerged as the molecular hub connecting LKB1 and PD-L1, by which Skp2 catalyzed K63-linked polyubiquitination on K136 and K280 residues to stabilize PD-L1 protein. Inhibition of Skp2 expression by short hairpin RNA or its E3 ligase activity by compound #25 abrogated intact expression of PD-L1 in vitro and generated a T cell-excluded "cold" TME in vivo. Thus, the LKB1-Skp2-PD-L1 regulatory loop was crucial for retaining PD-L1 protein expression and manipulation of this pathway would be a feasible approach for TME remodeling.
Conclusion: LKB1 and Skp2 are required for intact PD-L1 protein expression and TME remodeling in NSCLC. Inhibition of Skp2 resulted in a conversion from "hot" TME to "cold" TME and abrogated therapeutic outcomes of immunotherapy. Screening LKB1 and Skp2 status would be helpful to select recipients who may benefit from anti-PD-1/PD-L1 immunotherapy.
Keywords: Immune Checkpoint Inhibitor; Immunotherapy; Lung Cancer; Next generation sequencing - NGS; Tumor infiltrating lymphocyte - TIL.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: WL is an employee of Liaoning Kanghui Biotechnology. WL analyzed the next generation sequencing data and did not have access to interpret the bioinformatic and experimental results. The remaining authors did not have any financial association with Liaoning Kanghui Biotechnology or other biomedical companies.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
