Synthetic plasma pool cohort correction for affinity-based proteomics datasets allows multiple study comparison
- PMID: 39694815
- PMCID: PMC11653412
- DOI: 10.1093/bib/bbae657
Synthetic plasma pool cohort correction for affinity-based proteomics datasets allows multiple study comparison
Erratum in
-
Correction to: Synthetic plasma pool cohort correction for affinity-based proteomics datasets allows multiple study comparison.Brief Bioinform. 2024 Nov 22;26(1):bbaf112. doi: 10.1093/bib/bbaf112. Brief Bioinform. 2024. PMID: 40036723 Free PMC article. No abstract available.
Abstract
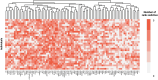
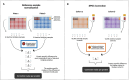
Proteomics stands as the crucial link between genomics and human diseases. Quantitative proteomics provides detailed insights into protein levels, enabling differentiation between distinct phenotypes. OLINK, a biotechnology company from Uppsala, Sweden, offers a targeted, affinity-based protein measurement method called Target 96, which has become prominent in the field of proteomics. The SCALLOP consortium, for instance, contains data from over 70.000 individuals across 45 independent cohort studies, all sampled by OLINK. However, when independent cohorts want to collaborate and quantitatively compare their target 96 protein values, it is currently advised to include 'identical biological bridging' samples in each sampling run to perform a reference sample normalization, correcting technical variations across measurements. Such a 'biological bridging sample' approach requires each of the involved cohorts to resend their biological bridging samples to OLINK to run them all together, which is logistically challenging, costly and time-consuming. Hence alternatives are searched and an evaluation of the current state of the art exposes the need for a more robust method that allows all OLINK Target 96 studies to compare proteomics data accurately and cost-efficiently. To meet these goals we developed the Synthetic Plasma Pool Cohort Correction, the 'SPOC correction' approach, based on the use of an OLINK-composed synthetic plasma sample. The method can easily be implemented in a federated data-sharing context which is illustrated on a sepsis use case.
Keywords: biomarkers; normalization; protein quantification; proteomics.
© The Author(s) 2024. Published by Oxford University Press.
Figures





References
-
- Proximity T, Assay E, Ab OP. et al. White Paper PEA – A High-Multiplex Immunoassay Technology with qPCR or NGS Readout. Olink proteomics AB. https://olink.com/.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

