Leveraging artificial intelligence and machine learning to accelerate discovery of disease-modifying therapies in type 1 diabetes
- PMID: 39694914
- PMCID: PMC11832708
- DOI: 10.1007/s00125-024-06339-6
Leveraging artificial intelligence and machine learning to accelerate discovery of disease-modifying therapies in type 1 diabetes
Abstract
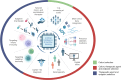
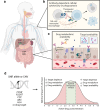
Progress in developing therapies for the maintenance of endogenous insulin secretion in, or the prevention of, type 1 diabetes has been hindered by limited animal models, the length and cost of clinical trials, difficulties in identifying individuals who will progress faster to a clinical diagnosis of type 1 diabetes, and heterogeneous clinical responses in intervention trials. Classic placebo-controlled intervention trials often include monotherapies, broad participant populations and extended follow-up periods focused on clinical endpoints. While this approach remains the 'gold standard' of clinical research, efforts are underway to implement new approaches harnessing the power of artificial intelligence and machine learning to accelerate drug discovery and efficacy testing. Here, we review emerging approaches for repurposing agents used to treat diseases that share pathogenic pathways with type 1 diabetes and selecting synergistic combinations of drugs to maximise therapeutic efficacy. We discuss how emerging multi-omics technologies, including analysis of antigen processing and presentation to adaptive immune cells, may lead to the discovery of novel biomarkers and subsequent translation into antigen-specific immunotherapies. We also discuss the potential for using artificial intelligence to create 'digital twin' models that enable rapid in silico testing of personalised agents as well as dose determination. To conclude, we discuss some limitations of artificial intelligence and machine learning, including issues pertaining to model interpretability and bias, as well as the continued need for validation studies via confirmatory intervention trials.
Keywords: Artificial intelligence; Digital twin; Drug discovery; Drug repurposing; Drug response; Immunotherapy; Machine learning; Pharmacogenetics; Precision medicine; Review; Type 1 diabetes.
© 2024. The Author(s).
Conflict of interest statement
Acknowledgements: The authors would like to thank the NIH Type 1 Diabetes TrialNet, Immune Tolerance Network and the Wanek Family Project Clinical Trial Group investigators for helpful discussions and/or published clinical trial data referenced throughout this manuscript, particularly in Table 2 and Fig. 2a. Funding: Work in the authors’ laboratories is supported by NIH P01 AI042288 (to TMB) and K99 DK140511 (to MRS); The Leona M. and Harry B. Helmsley Charitable Trust 2019PG-T1D011 (to TMB); Breakthrough T1D (formerly JDRF) Postdoctoral Fellowship 3-PDF-2022-1137-A-N (to MRS); Orlando Brown, Jr (to MAC); and the Emilie Rosebud Diabetes Research Foundation (to MAC). The funders of this work were not involved in designing this review; collecting, analysing or interpreting data; or writing this report. The funders did not impose any restrictions regarding the publication of this report. Authors’ relationships and activities: MAC has received research support from Dexcom and Abbott Diabetes Care and consulting fees from Glooko. The authors declare that there are no other relationships or activities that might bias, or be perceived to bias, their work. Contribution statement: All authors were responsible for drafting the article and reviewing it critically for important intellectual content. All authors approved the version to be published.
Figures

References
-
- OECD (2023) Artificial intelligence in science: challenges, opportunities and the future of research. OECD Publishing, Paris. 10.1787/a8d820bd-en
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

