DARPins as a novel tool to detect and degrade p73
- PMID: 39695090
- PMCID: PMC11655841
- DOI: 10.1038/s41419-024-07304-2
DARPins as a novel tool to detect and degrade p73
Abstract
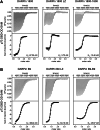
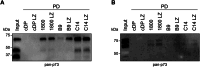
The concept of Targeted Protein Degradation (TPD) has been introduced as an attractive alternative to the development of classical inhibitors. TPD can extend the range of proteins that can be pharmacologically targeted beyond the classical targets for small molecule inhibitors, as a binding pocket is required but its occupancy does not need to lead to inhibition. The method is based on either small molecules that simultaneously bind to a protein of interest and to a cellular E3 ligase and bring them in close proximity (molecular glue) or a bi-functional molecule synthesized from the chemical linkage of a target protein-specific small molecule and one that binds to an E3 ligase (Proteolysis Targeting Chimeras (PROTAC)). The further extension of this approach to bioPROTACs, in which a small protein-based binding module is fused directly to an E3 ligase or an E3 ligase adaptor protein, makes virtually all proteins amenable to targeted degradation, as this method eliminates the requirement for binding pockets for small molecules. Designed Ankyrin Repeat Proteins (DARPins) represent a very attractive class of small protein-based binding modules that can be used for the development of bioPTOTACS. Here we describe the characterization of two DARPins generated against the oligomerization domain and the SAM domain of the transcription factor p73, a member of the p53 protein family. The DARPins can be used for (isoform-)selective pulldown experiments both in cell culture as well as primary tissue lysates. We also demonstrate that they can be used for staining in cell culture experiments. Fusing them to the speckle type POZ protein (SPOP), an adaptor protein for cullin-3 E3 ligase complexes, yields highly selective and effective degraders. We demonstrate that selective degradation of the ΔNp73α isoform reactivates p53.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: Andreas Plückthun is a cofounder and shareholder of Molecular Partners AG, who are commercializing the DARPin technology. All other authors declare that they have no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

