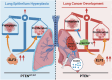
Overexpression of ELF3 in the PTEN-deficient lung epithelium promotes lung cancer development by inhibiting ferroptosis
- PMID: 39695109
- PMCID: PMC11655876
- DOI: 10.1038/s41419-024-07274-5
Overexpression of ELF3 in the PTEN-deficient lung epithelium promotes lung cancer development by inhibiting ferroptosis
Erratum in
-
Correction: Overexpression of ELF3 in the PTEN-deficient lung epithelium promotes lung cancer development by inhibiting ferroptosis.Cell Death Dis. 2025 Feb 10;16(1):85. doi: 10.1038/s41419-025-07397-3. Cell Death Dis. 2025. PMID: 39929811 Free PMC article. No abstract available.
Abstract
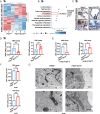
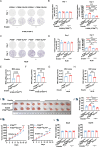
Ferroptosis has been shown to play a crucial role in preventing cancer development, but the underlying mechanisms of dysregulated genes and genetic alternations driving cancer development by regulating ferroptosis remain unclear. Here, we showed that the synergistic role of ELF3 overexpression and PTEN deficiency in driving lung cancer development was highly dependent on the regulation of ferroptosis. Human ELF3 (hELF3) overexpression in murine lung epithelial cells only caused hyperplasia with increased proliferation and ferroptosis. hELF3 overexpression and Pten genetic disruption significantly induced lung tumor development with increased proliferation and inhibited ferroptosis. Mechanistically, we found it was due to the induction of SCL7A11, a typical ferroptosis inhibitor, and ELF3 directly and positively regulated SCL7A11 in the PTEN-deficient background. Erastin-mediated inhibition of SCL7A11 induced ferroptosis in cells with ELF3 overexpression and PTEN deficiency and thus inhibited cell colony formation and tumor development. Clinically, human lung tumors showed a negative correlation between ELF3 and PTEN expression and a positive correlation between ELF3 and SCL7A11 in a subset of human lung tumors with PTEN-low expression. ELF3 and SCL7A11 expression levels were negatively associated with lung cancer patients' survival rates. In summary, ferroptosis induction can effectively attenuate lung tumor development induced by ELF3 overexpression and PTEN downregulation or loss-of-function mutations.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: All methods were performed in accordance with the relevant guidelines and regulations. All animal experiments were approved by the Biomedical Research Ethics Committee, Zhejiang University, China. Reference Number: 13888. Consent to publish: Informed consent was obtained from all participants.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

