Collagen signaling and matrix stiffness regulate multipotency in glandular epithelial stem cells in mice
- PMID: 39695111
- PMCID: PMC11655882
- DOI: 10.1038/s41467-024-54843-5
Collagen signaling and matrix stiffness regulate multipotency in glandular epithelial stem cells in mice
Abstract
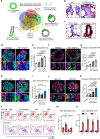
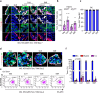
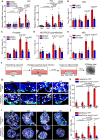
Glandular epithelia, including mammary gland (MG) and prostate, are composed of luminal and basal cells. During embryonic development, glandular epithelia arise from multipotent stem cells (SCs) that are replaced after birth by unipotent basal and unipotent luminal SCs. Different conditions, such as basal cell transplantation, luminal cell ablation, and oncogene expression can reinduce adult basal SC (BaSCs) multipotency in different glandular epithelia. The mechanisms regulating the reactivation of multipotency are incompletely understood. Here, we have found that Collagen I expression is commonly upregulated in BaSCs across the different multipotent conditions. Increasing collagen concentration or stiffness of the extracellular matrix (ECM) promotes BaSC multipotency in MG and prostate organoids. Single cell RNA-seq of MG organoids in stiff conditions have uncovered the importance of β1 integrin/FAK/AP-1 axis in the regulation of BaSC multipotency. Altogether our study uncovers the key role of Collagen signaling and ECM stiffness in the regulation of multipotency in glandular epithelia.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Lloyd-Lewis, B., Harris, O. B., Watson, C. J. & Davis, F. M. Mammary stem cells: premise, properties, and perspectives. Trends Cell Biol.27, 556–567 (2017). - PubMed
-
- Ousset, M. et al. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat. Cell Biol.14, 1131–1138 (2012). - PubMed
-
- Van Keymeulen, A. et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature479, 189–193 (2011). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

