Single-cell transcriptomic and spatial analysis reveal the immunosuppressive microenvironment in relapsed/refractory angioimmunoblastic T-cell lymphoma
- PMID: 39695118
- PMCID: PMC11655871
- DOI: 10.1038/s41408-024-01199-0
Single-cell transcriptomic and spatial analysis reveal the immunosuppressive microenvironment in relapsed/refractory angioimmunoblastic T-cell lymphoma
Abstract
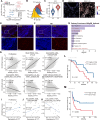
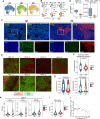
Angioimmunoblastic T-cell lymphoma (AITL) is a kind of aggressive T-cell lymphoma with significant enrichment of non-malignant tumor microenvironment (TME) cells. However, the complexity of TME in AITL progression is poorly understood. We performed single-cell RNA-Seq (scRNA-seq) and imaging mass cytometry (IMC) analysis to compare the cellular composition and spatial architecture between relapsed/refractory AITL (RR-AITL) and newly diagnosed AITL (ND-AITL). Our results showed that the malignant T follicular helper (Tfh) cells showed significantly increased proliferation driven by transcriptional activation of YY1 in RR-AITL, which is markedly associated with the poor prognosis of AITL patients. The CD8+ T cell proportion and cytotoxicity decreased in RR-AITL TME, resulting from elevated expression of the inhibitory checkpoints such as PD-1, TIGIT, and CTLA4. Notably, the transcriptional pattern of B cells in RR-AITL showed an intermediate state of malignant transformation to B-cell-lymphoma, and contributed to immune evasion by highly expressing CD47 and PD-L1. Besides, compared to ND-AITL samples, myeloid-cells-centered spatial communities were more prevalent but showed reduced phagocytic activity and impaired antigen processing and presentation in RR-AITL TME. Furthermore, specific inhibitory ligand-receptor interactions, such as CLEC2D-KLRB1, CTLA4-CD86, and MIF-CD74, were exclusively identified in the RR-AITL TME. Our study provides a high-resolution characterization of the immunosuppression ecosystem and reveals the potential therapeutic targets for RR-AITL patients.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: The study was reviewed and approved by the Ethical Committee of the Nanjing Medical University (Nanjing, Jiangsu, China). The study was performed after receiving written informed consent from patients, by the principles outlined in the Declaration of Helsinki.
Figures





References
-
- Nguyen TB, Sakata-Yanagimoto M, Fujisawa M, Nuhat ST, Miyoshi H, Nannya Y, et al. Dasatinib is an effective treatment for angioimmunoblastic T-cell lymphoma. Cancer Res. 2020;80:1875–84. 10.1158/0008-5472.CAN-19-2787. - PubMed
-
- Dunleavy K, Wilson WH, Jaffe ES. Angioimmunoblastic T cell lymphoma: pathobiological insights and clinical implications. Curr Opin Hematol. 2007;14:348–53. 10.1097/MOH.0b013e328186ffbf. - PubMed
Publication types
MeSH terms
Grants and funding
- 32200590/National Natural Science Foundation of China (National Science Foundation of China)
- 82370193/National Natural Science Foundation of China (National Science Foundation of China)
- 81972358/National Natural Science Foundation of China (National Science Foundation of China)
- 91959113/National Natural Science Foundation of China (National Science Foundation of China)
- 82372897/National Natural Science Foundation of China (National Science Foundation of China)
- 82170185/National Natural Science Foundation of China (National Science Foundation of China)
- BK20210530/Natural Science Foundation of Jiangsu Province (Jiangsu Provincial Natural Science Foundation)
- BK20211376/Natural Science Foundation of Jiangsu Province (Jiangsu Provincial Natural Science Foundation)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

