SMURF1 and SMURF2 directly target GLI1 for ubiquitination and proteasome-dependent degradation
- PMID: 39695131
- PMCID: PMC11655642
- DOI: 10.1038/s41420-024-02260-4
SMURF1 and SMURF2 directly target GLI1 for ubiquitination and proteasome-dependent degradation
Abstract
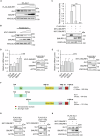
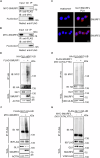
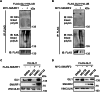
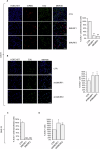
The transcription factor GLI1 is the main and final effector of the Hedgehog signaling pathway, which is involved in embryonic development, cell proliferation and stemness. Whether activated through canonical or non-canonical mechanisms, GLI1 aberrant activity is associated with Hedgehog-dependent cancers, including medulloblastoma, as well as other tumoral contexts. Notwithstanding a growing body of evidence, which have highlighted the potential role of post translational modifications of GLI1, the complex mechanisms modulating GLI1 stability and activity have not been fully elucidated. Here, we present a novel role played by SMURF1 and SMURF2 in the suppression of the Hedgehog/GLI signaling pathway through a direct targeting of GLI1. Indeed, the two SMURFs can interact with GLI1, exploiting the proline rich regions present on GLI1 protein, and trigger its polyubiquitination and proteasomal degradation, leading to a suppression of the Hedgehog pathway activity and a reduction of Hh-dependent tumor cell proliferation. Overall, this study adds new relevance to a tumor suppressive role of SMURFs on the Hedgehog pathway and confers upon them the status of potential therapeutic tools, either in canonical or non-canonical Hedgehog pathway aberrant activation.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: The experiments performed in this work did not use material (i.e., human or mice samples) that requires ethical approval.
Figures


References
-
- Zhang Y, Beachy PA. Cellular and molecular mechanisms of Hedgehog signalling. Nat Rev Mol Cell Biol. 2023;24:668–87. - PubMed
Grants and funding
- RG1221816C0C91FE/Sapienza Università di Roma (Sapienza University of Rome)
- AR1221816BA289E7/Sapienza Università di Roma (Sapienza University of Rome)
- AR12117A81C83875/Sapienza Università di Roma (Sapienza University of Rome)
- AR223188B0F7C930/Sapienza Università di Roma (Sapienza University of Rome)
- n/A/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
LinkOut - more resources
Full Text Sources
Medical

