Targeting PPARγ via SIAH1/2-mediated ubiquitin-proteasomal degradation as a new therapeutic approach in luminal-type bladder cancer
- PMID: 39695138
- PMCID: PMC11655661
- DOI: 10.1038/s41419-024-07298-x
Targeting PPARγ via SIAH1/2-mediated ubiquitin-proteasomal degradation as a new therapeutic approach in luminal-type bladder cancer
Abstract
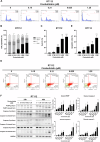
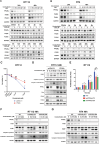
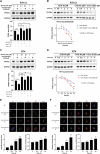
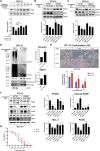
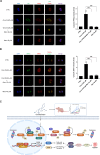
Bladder cancer (BC) is the second most prevalent genitourinary malignancy worldwide. Despite recent approvals of immune checkpoint inhibitors and targeted therapy for muscle invasive or recurrent BC, options remain limited for patients with non-muscle invasive BC (NMIBC) refractory to Bacillus Calmette-Guérin (BCG) and chemotherapy. NMIBC is more frequently classified as a luminal subtype, in which increased PPARγ activity is a key feature in promoting tumor growth and evasion of immunosurveillance. Cinobufotalin is one of the major compound of bufadienolides, the primary active components of toad venom that has been utilized in the clinical treatment of cancer. We herein focused on cinobufotalin, examining its anticancer activity and molecular mechanisms in luminal-type NMIBC. Our results newly reveal that cinobufotalin strongly suppresses the viability and proliferation of luminal BC cells with minimal cytotoxic effects on normal uroepithelial cells, and exhibits significant antitumor activity in a RT112 xenograft BC model. Mechanistically, our sub-G1-phase cell accumulation, Annexin V staining, caspase-3/8/9 activation, and PARP activation analyses show that cinobufotalin induces apoptosis in luminal-type BC cells. Cinobufotalin significantly inhibited the levels of PPARγ and its downstream targets, as well as lipid droplet formation and free fatty acid levels in RT112 cells. PPARγ overexpression rescued RT112 cells from cinobufotalin-induced apoptosis and mitigated the downregulation of FASN and PLIN4. Finally, we show seemingly for the first time that cinobufotalin promotes SIAH1/2-mediated proteasomal degradation of PPARγ in luminal BC cells. Together, these findings compellingly support the idea that cinobufotalin could be developed as a promising therapeutic agent for treating luminal-type NMIBC.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethical approval: All animal experiments followed the ethical guidelines, and the protocols have been reviewed and approved by the institutional Animal Care and Use Committee of Taipei Medical University (IACUC approval no: LAC-2021-0471).
Figures


References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Babjuk M, Burger M, Zigeuner R, Shariat SF, van Rhijn BW, Comperat E. et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: update 2013. Eur Urol. 2013;64:639–53. 10.1016/j.eururo.2013.06.003. - PubMed
-
- Joyce DD, Sharma V, Williams SB. Cost-effectiveness and economic impact of bladder cancer management: an updated review of the literature. Pharmacoeconomics. 2023;41:751–69. 10.1007/s40273-023-01273-8. - PubMed
-
- Balasubramanian A, Gunjur A, Weickhardt A, Papa N, Bolton D, Lawrentschuk N. et al. Adjuvant therapies for non-muscle-invasive bladder cancer: advances during BCG shortage. World J Urol. 2022;40:1111–24. 10.1007/s00345-021-03908-x. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

