Design of pseudosymmetric protein hetero-oligomers
- PMID: 39695145
- PMCID: PMC11655659
- DOI: 10.1038/s41467-024-54913-8
Design of pseudosymmetric protein hetero-oligomers
Abstract
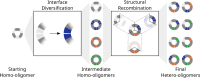
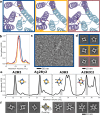
Pseudosymmetric hetero-oligomers with three or more unique subunits with overall structural (but not sequence) symmetry play key roles in biology, and systematic approaches for generating such proteins de novo would provide new routes to controlling cell signaling and designing complex protein materials. However, the de novo design of protein hetero-oligomers with three or more distinct chains with nearly identical structures is a challenging unsolved problem because it requires the accurate design of multiple protein-protein interfaces simultaneously. Here, we describe a divide-and-conquer approach that breaks the multiple-interface design challenge into a set of more tractable symmetric single-interface redesign tasks, followed by structural recombination of the validated homo-oligomers into pseudosymmetric hetero-oligomers. Starting from de novo designed circular homo-oligomers composed of 9 or 24 tandemly repeated units, we redesigned the inter-subunit interfaces to generate 19 new homo-oligomers and structurally recombined them to make 24 new hetero-oligomers, including ABC heterotrimers, A2B2 heterotetramers, and A3B3 and A2B2C2 heterohexamers which assemble with high structural specificity. The symmetric homo-oligomers and pseudosymmetric hetero-oligomers generated for each system have identical or nearly identical backbones, and hence are ideal building blocks for generating and functionalizing larger symmetric and pseudosymmetric assemblies.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: A provisional patent application has been filed (63/486,872) by the University of Washington, listing R.D.K., B.I.M.W., B.L.S., S.L., and D.B. as inventors or contributors. The remaining authors declare no competing interests.
Figures



Update of
-
Stepwise design of pseudosymmetric protein hetero-oligomers.bioRxiv [Preprint]. 2023 Apr 7:2023.04.07.535760. doi: 10.1101/2023.04.07.535760. bioRxiv. 2023. Update in: Nat Commun. 2024 Dec 18;15(1):10684. doi: 10.1038/s41467-024-54913-8. PMID: 37066191 Free PMC article. Updated. Preprint.
References
-
- Levy, E. D. & Teichmann, S. A. Chapter two - structural, evolutionary, and assembly principles of protein oligomerization. In Oligomerization in Health and Disease (eds. Giraldo, J. & Ciruela, F.) 25–51 (Academic Press, 2013). - PubMed
-
- Lang, D., Thoma, R., Henn-Sax, M., Sterner, R. & Wilmanns, M. Structural evidence for evolution of the β/α barrel scaffold by gene duplication and fusion. Science289, 1546–1550 (2000). - PubMed
-
- Khusial, P., Plaag, R. & Zieve, G. W. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem. Sci.30, 522–528 (2005). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- P30 GM124169/GM/NIGMS NIH HHS/United States
- R01 GM105691/GM/NIGMS NIH HHS/United States
- R01 GM139752/GM/NIGMS NIH HHS/United States
- R35 GM148166/GM/NIGMS NIH HHS/United States
- ALTF 139-2018/European Molecular Biology Organization (EMBO)
- S10 OD021832/OD/NIH HHS/United States
- HHSN272201700059C/AI/NIAID NIH HHS/United States
- P41 GM128577/GM/NIGMS NIH HHS/United States
- shared instrumentation grant S10OD021832/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- RM1 GM149374/GM/NIGMS NIH HHS/United States
- #OPP1156262/Bill and Melinda Gates Foundation (Bill & Melinda Gates Foundation)
- DGE-1762114/National Science Foundation (NSF)
- INV-010680/GATES/Gates Foundation/United States
- S10 OD018483/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources

