Salmonella Typhimurium effector SseI regulates host peroxisomal dynamics to acquire lysosomal cholesterol
- PMID: 39695325
- PMCID: PMC11811301
- DOI: 10.1038/s44319-024-00328-x
Salmonella Typhimurium effector SseI regulates host peroxisomal dynamics to acquire lysosomal cholesterol
Abstract
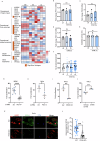
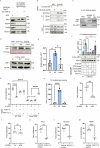
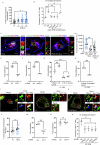
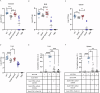
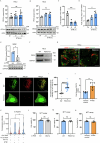
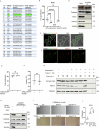
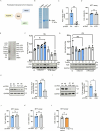
Salmonella enterica serotype Typhimurium (Salmonella) resides and multiplies intracellularly in cholesterol-rich compartments called Salmonella-containing vacuoles (SCVs) with actin-rich tubular extensions known as Salmonella-induced filaments (SIFs). SCV maturation depends on host-derived cholesterol, but the transport mechanism of low-density lipoprotein (LDL)-derived cholesterol to SCVs remains unclear. Here we find that peroxisomes are recruited to SCVs and function as pro-bacterial organelle. The Salmonella effector protein SseI is required for the interaction between peroxisomes and the SCV. SseI contains a variant of the PTS1 peroxisome-targeting sequence, GKM, localizes to the peroxisomes and activates the host Ras GTPase, ADP-ribosylation factor-1 (ARF-1). Activation of ARF-1 leads to the recruitment of phosphatidylinsolitol-5-phosphate-4 kinase and the generation of phosphatidylinsolitol-4-5-bisphosphate on peroxisomes. This enhances the interaction of peroxisomes with lysosomes and allows for the transfer of lysosomal cholesterol to SCVs using peroxisomes as a bridge. Salmonella infection of peroxisome-depleted cells leads to the depletion of cholesterol on the SCVs, resulting in reduced SIF formation and bacterial proliferation. Taken together, our work identified peroxisomes as a target of Salmonella secretory effectors, and as conveyance of host cholesterol to enhance SCV stability, SIF integrity, and intracellular bacterial growth.
Keywords: Salmonella Typhimurium; ARF1 Activation; Cholesterol; Peroxisome-targeting Sequence; Peroxisomes.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- AD HOC (ORTHO) 2022-NCD-1/Indian Council of Medical Research (ICMR)
- MLP2105,HCP0047,Director-CDRI/Council of Scientific and Industrial Research, India (CSIR)
- WT_/Wellcome Trust/United Kingdom
- IA/E/21/1/506319/Wellcome Trust DBT India Alliance (India Alliance)
- PDF/2021/002843 SRG/2019/000268/DST | Science and Engineering Research Board (SERB)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

