Reliability of wing morphometrics for species identification of human-biting black flies (Diptera: Simuliidae) in Thailand
- PMID: 39695748
- PMCID: PMC11658325
- DOI: 10.1186/s13071-024-06597-8
Reliability of wing morphometrics for species identification of human-biting black flies (Diptera: Simuliidae) in Thailand
Abstract
Background: Fast and reliable species identification of black flies is essential for research proposes and effective vector control. Besides traditional identification based on morphology, which is usually supplemented with molecular methods, geometric morphometrics (GM) has emerged as a promising tool for identification. Despite its potential, no specific GM techniques have been established for the identification of black fly species.
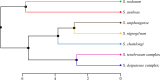
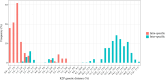
Methods: Adult female black flies collected using human bait, as well as those reared from pupae, were used in this study. Here, landmark-based GM analysis of wings was assessed for the first time to identify human-biting black fly species in Thailand, comparing this approach with the standard morphological identification method and DNA barcoding based on the mitochondrial cytochrome c oxidase subunit I (COI) gene. To explore genetic relationships between species, maximum likelihood (ML) and neighbor-joining (NJ) phylogenetic trees were built. Additionally, three different methods of species delimitation, i.e., assemble species by automatic partitioning (ASAP), generalized mixed yule coalescent (GMYC), and single Poisson tree processes (PTP), were utilized to identify the morphologically defined species. The effectiveness of a COI barcode in identifying black fly species was further examined through the best match (BM) and best close match (BCM) methods.
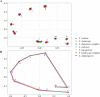
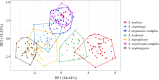
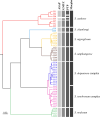
Results: Seven black fly species, namely Simulium tenebrosum Takaoka, Srisuka & Saeung, 2018 (complex), S. doipuiense Takaoka & Choochote, 2005 (complex), S. nigrogilvum Summers, 1911, S. nodosum Puri, 1933, S. asakoae Takaoka & Davies, 1995, S. chamlongi Takaoka & Suzuki, 1984, and S. umphangense Takaoka, Srisuka & Saeung, 2017 were morphologically identified. Compared with the standard method, the GM analysis based on wing shape showed high success in separating species, achieving an overall accuracy rate of 88.54%. On the other hand, DNA barcoding surpassed wing GM for species identification with a correct identification rate of 98.57%. Species delimitation analyses confirmed the validity of most nominal species, with an exception for S. tenebrosum complex and S. doipuiense complex, being delimited as a single species. Moreover, the analyses unveiled hidden diversity within S. asakoae, indicating the possible existence of up to four putative species.
Conclusions: This study highlights the potential of wing GM as a promising and reliable complementary tool for species identification of human-biting black flies in Thailand.
Keywords: Simulium; DNA barcodes; Hematophagous insect; Medical entomology; Morphometric analysis; Species delimitation.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The procedures and research methodology used in this study were approved by the Research Ethics Committee (Institutional Animal Care and Use Committee) (protocol number: 28/2564) of the Faculty of Medicine, Chiang Mai University, Chiang Mai Province, Thailand. Consent for publication: All authors have read and approved the final version of the manuscript. Competing interests: The authors declare no competing interests.
Figures




References
-
- Adler PH. World blackflies (Diptera: Simuliidae): a comprehensive revision of the taxonomic and geographical inventory [2024]. 2024. https://biomia.sites.clemson.edu/pdfs/blackflyinventory.pdf.
-
- Schaffartzik A, Marti E, Crameri R, Rhyner C. Cloning, production and characterization of antigen 5 like proteins from Simuliumvittatum and Culicoidesnubeculosus, the first cross-reactive allergen associated with equine insect bite hypersensitivity. Vet Immunol Immunopathol. 2010;137:76–83. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

