Advancing diagnosis and early risk assessment of preeclampsia through noninvasive cell-free DNA methylation profiling
- PMID: 39695764
- PMCID: PMC11656954
- DOI: 10.1186/s13148-024-01798-5
Advancing diagnosis and early risk assessment of preeclampsia through noninvasive cell-free DNA methylation profiling
Abstract
Background: Aberrant embryo implantation and suboptimal placentation can lead to (severe) complications such as preeclampsia and fetal growth restriction later in pregnancy. Current identification of high-risk pregnancies relies on a combination of risk factors, biomarkers, and ultrasound examinations, a relatively inaccurate approach. Previously, aberrant DNA methylation due to placental hypoxia has been identified as a potential marker of placental insufficiency and, hence, potential (future) pregnancy complications. The goal of the Early Prediction of prEgnancy Complications Testing, or the ExPECT study, is to validate a genome-wide, cell-free DNA (cfDNA) methylation strategy to diagnose preeclampsia accurately. More importantly, the predictive potential of this strategy is also explored to reliably identify high-risk pregnancies early in gestation. Furthermore, a longitudinal study was conducted, including sequential blood samples from pregnant individuals experiencing both uneventful and complicated gestations, to assess the methylation dynamics of cfDNA throughout these pregnancies. A significant strength of this study is its enzymatic digest, which enriches CpG-rich regions across the genome without the need for proprietary reagents or prior selection of regions of interest. This makes it useful for the cost-effective discovery of novel markers.
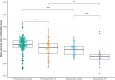
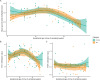
Results: Investigation of methylation patterns throughout pregnancy showed different methylation trends between unaffected and affected pregnancies. We detected differentially methylated regions (DMRs) in pregnancies complicated with preeclampsia as early as 12 weeks of gestation, with distinct differences in the methylation profile between early and late pregnancy. Two classification models were developed to diagnose and predict preeclampsia, demonstrating promising results on a small set of validation samples.
Conclusions: This study offers valuable insights into methylation changes at specific genomic regions throughout pregnancy, revealing critical differences between normal and complicated pregnancies. The power of noninvasive cfDNA methylation profiling was successfully proven, suggesting the potential to integrate this noninvasive approach into routine prenatal care.
Keywords: Cell-free DNA; CfDNA methylation; DMRs; Differentially methylated regions; Epigenetics; Placental insufficiency; Prediction; Preeclampsia; Prenatal.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The Ethical Committee of the University Hospital Ghent approved all research included in this study (2019/0056). Written informed consent was obtained from all participants under the approval of the Ethical Committee. Consent for publication: Applicable. Competing interests: All authors declare that they have no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

