Patterns of genomic instability in > 2000 patients with ovarian cancer across six clinical trials evaluating olaparib
- PMID: 39695768
- PMCID: PMC11657106
- DOI: 10.1186/s13073-024-01413-5
Patterns of genomic instability in > 2000 patients with ovarian cancer across six clinical trials evaluating olaparib
Abstract
Background: The introduction of poly(ADP-ribose) polymerase (PARP) inhibitors represented a paradigm shift in the treatment of ovarian cancer. Genomic data from patients with high-grade ovarian cancer in six phase II/III trials involving the PARP inhibitor olaparib were analyzed to better understand patterns and potential causes of genomic instability.
Patients and methods: Homologous recombination deficiency (HRD) was assessed in 2147 tumor samples from SOLO1, PAOLA-1, Study 19, SOLO2, OPINION, and LIGHT using next-generation sequencing technology. Genomic instability scores (GIS) were assessed in BRCA1 and/or BRCA2 (BRCA)-mutated (BRCAm), non-BRCA homologous recombination repair-mutated (non-BRCA HRRm), and non-HRRm tumors.
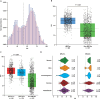
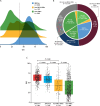
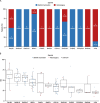
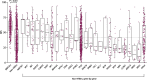
Results: BRCAm was identified in 1021/2147 (47.6%) tumors. BRCAm tumors had significantly higher GIS than non-BRCAm tumors (P < 0.001) and high biallelic loss (815/838; 97.3%) regardless of germline (658/672; 97.9%) or somatic (101/108; 93.5%) BRCAm status. In non-BRCA HRRm tumors (n = 121) a similar proportion were HRD-positive (GIS ≥ 42: 55/121; 45.5%) relative to HRD-negative (GIS < 42: 52/121; 43.0%). GIS was highly variable in non-BRCA HRRm (median 42 [interquartile range (IQR) 29-58]) and non-HRRm (n = 1005; median 32 [IQR 20-55]) tumors. Gene mutations with high GIS included HRR genes BRIP1 (median 46 [IQR 41-58]), RAD51C (median 58 [IQR 48-66]), RAD51D (median 62 [IQR 54-69]), and PALB2 (median 64 [IQR 58-74]), and non-HRR genes NF1 (median 49 [IQR 25-60]) and RB1 (median 55 [IQR 30-71]). CCNE1-amplified and PIK3CA-mutated tumors had low GIS (CCNE1-amplified: median 24 [IQR 18-29]; PIK3CA-mutated: median 32 [IQR 14-52]) and were predominantly non-BRCAm.
Conclusions: These analyses provide valuable insight into patterns of genomic instability and potential drivers of HRD, besides BRCAm, in ovarian cancer and will help guide future research into the potential clinical effectiveness of anti-cancer treatments in ovarian cancer, including PARP inhibitors as well as other precision oncology agents.
Trial registration: The SOLO1 trial was registered at ClinicalTrials.gov (NCT01844986) on April 30, 2013; the PAOLA-1 trial was registered at ClinicalTrials.gov (NCT02477644) on June 18, 2015 (retrospectively registered); Study 19 was registered at ClinicalTrials.gov (NCT00753545) on September 12, 2008 (retrospectively registered); the SOLO2 trial was registered at ClinicalTrials.gov (NCT01874353) on June 7, 2013; the OPINION trial was registered at ClinicalTrials.gov (NCT03402841) on January 3, 2018; the LIGHT trial was registered at ClinicalTrials.gov (NCT02983799) on November 4, 2016.
Keywords: Genomic instability; Olaparib; Ovarian cancer; Translational research.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All trials and analyses were performed in accordance with the principles of the Declaration of Helsinki and Good Clinical Practice guidelines and were approved by the appropriate Institutional Review Boards. All patients provided written informed consent. Consent for publication: Not applicable. Competing interests: AB reports full-time employment with AstraZeneca during the conduct of the study and AstraZeneca stock ownership. IR-C reports grants to self from BMS, MSD, and Roche; grants to their institution from AstraZeneca, BMS, Merck Serono, MSD, Novartis, and Roche; consulting fees from AstraZeneca, Agenus, Advaxis, Amgen, BMS, Clovis, Deciphera, Genmab, GSK, Mersana, Merck Serono, MSD, Novartis, Pfizer, PharmaMar, Roche/Genentech, and Tesaro; payment or honoraria to self for lectures, presentations, speakers' bureaus, manuscript writing, or educational events from AbbVie, Agenus, Advaxis, Amgen, AstraZeneca, BMS, Clovis, Deciphera, Genmab, GSK, Mersana, Merck Serono, MSD, Novartis, Pfizer, PharmaMar, Roche, and Tesaro; payment or honoraria to institution for lectures, presentations, speakers' bureaus, manuscript writing, or educational events from BMS, GSK, MSD, and Roche; and support for attending meetings and/or travel from AstraZeneca, GSK, and Roche. ER reports presentations for BMS, AstraZeneca, Clovis, Roche Diagnostic, GSK, and Roche; advisory boards for AstraZeneca, Roche, and GSK; and congress participation for BMS and AstraZeneca. KC reports grants to her institution from The Irish Cancer Society Clinician Research Leadership Award 2021, MSD Ireland, and ImmunoGen; consulting fees from GSK Ireland and Nextcure; payment or honoraria for lectures, presentations, speakers' bureaus, manuscript writing, or educational events from AstraZeneca, GSK Ireland, MJH Life Sciences, and MSD Ireland; payment for expert testimony from St Vincent’s Health; support for attending meetings from MSD Ireland, Pfizer, and Roche Ireland; participation on a data safety monitoring board or advisory board for AstraZeneca, Eisai, GSK Ireland, and Merck; voluntary board member for Arc Cancer Support Centers; and voluntary advisory role for National Cancer Control Programme Ireland and National Center for Pharmacoeconomics Ireland. FS reports participating on scientific advisory boards for AstraZeneca, Zentalis, and GlaxoSmithKline; and receiving grant/research support to their institution from Repare Therapeutics, AstraZeneca, and InstilBio. CA reports receiving advisory board fees from AbbVie, AstraZeneca/Merck, Eisai/Merck, Mersana Therapeutics, Repare Therapeutics, and Roche/Genentech; participation on an advisory board for Blueprint Medicine; participation on the board of directors for GOG Foundation and NRG Oncology; clinical trial funding to her institution from AstraZeneca for this study; and clinical trial funding to her institution from AbbVie, AstraZeneca, Clovis, and Genentech. AL reports grants from AstraZeneca and Sanofi; consulting fees from Seattle Genetics; honoraria/reimbursement and advisory board fees from AstraZeneca; advisory board fees or continuing medical education from Ability Pharma, Biocad, Clovis Oncology, GSK, Medscape, Merck Serono, MSD, TouchCongress, and Zentalis; support for attending meetings and/or travel from AstraZeneca, Clovis Oncology, GSK, and Roche; and participation on a data safety monitoring board or advisory board for ARIEL4 and TROPHIMMUNE. AP reports honoraria for advisory or speaker activities and/or congress travel support from Roche, AstraZeneca, Tesaro, GSK, MSD, and Clovis; and participation on a data safety monitoring board from MSD. SL reports support for the present manuscript from AstraZeneca; grants or contracts to their institution from Merck, AstraZeneca, Regeneron, Roche, Repare, GSK, and Seagen; consulting fees from Novocure, Merck, AstraZeneca, GSK, Eisai, and Shattuck Labs; payment or honoraria for lectures, presentations, speakers' bureaus, manuscript writing, or educational events from AstraZeneca, GSK, and Eisai; and participation on a data safety monitoring board or advisory board from AstraZeneca. EP-L reports lecture fees, fees for serving on a speakers' bureau, and travel support from AstraZeneca, GSK, Roche, and Tesaro; lecture fees from Clovis Oncology and Pfizer; expert testimony fees from AstraZeneca; support for attending meetings and/or travel from AstraZeneca and GSK; fees from AstraZeneca, Incyte, and Roche for participating on a data safety monitoring board or advisory board; and employment by ARCAGY Research. BY reports consulting or advisory roles for Amgen, AstraZeneca, Bayer, BMS, Clovis, ECS Progastrin, GSK, MSD, Myriad, Novartis, and Roche; and travel support from AstraZeneca, Bayer, MSD, and Roche. JL reports receiving research grants from AstraZeneca and Merck/MSD; lecture fees from Clovis Oncology, AstraZeneca, Neopharm, GSK, and MSD/Merck; and advisory board fees from AstraZeneca, GSK, Artios Pharma, Clovis Oncology, ImmunoGen, Mersana, Bristol Myers Squibb, Nuvation, Ellipses Pharma, VBL Therapeutics, Eisai, Sutro Bio, and Immagene, outside of the submitted work. UM reports participation in scientific advisory boards for Allarity, NextCure, Trillium, Agenus, ImmunoGen, Profound Bio, Eisai, Novartis, Boehringer Ingelheim, the Ovarian Cancer Research Alliance, MorphoSys, and CureLab; participation in a data safety monitoring board for Alkermes and Symphogen; consulting for Merck, GSK, and AstraZeneca; participation in a speakers' bureau from Med Learning Group; and funding from the Dana-Farber/Harvard Cancer Center Ovarian Cancer SPORE grant (P50CA240243), the Breast Cancer Research Fund, and the Dana-Farber/Harvard Cancer Center grant (2P30CA006516-57). CG reports clinical trial funding for this study to his institution from AstraZeneca; clinical research grants to his institution from Aprea, AstraZeneca, BergenBio, Clovis, GlaxoSmithKline, Medannexin, MSD, Novartis, Nucana, and Tesaro; personal consulting fees from AstraZeneca, GlaxoSmithKline, MSD, and Tesaro; honoraria for lectures/presentations from AstraZeneca, Chugai, Clovis Oncology, GlaxoSmithKline, MSD, Nucana, Roche, Takeda, and Tesaro; honoraria for lectures/presentations/preparing educational materials from Cor2Ed; advisory board attendance for AstraZeneca, Chugai, GlaxoSmithKline, MSD, Nucana, Roche, and Tesaro; and being a committee member on the Scottish Medicines Consortium. KMT reports salary and stock options from Myriad Genetics, Inc. ZL reports full-time employment with AstraZeneca and AstraZeneca stock ownership. DRH reports full-time employment with AstraZeneca and AstraZeneca stock ownership. CEE reports full-time employment with AstraZeneca and AstraZeneca stock ownership. SD reports full-time employment with AstraZeneca and AstraZeneca stock ownership. CE reports full-time employment with AstraZeneca and AstraZeneca stock ownership. PL-S reports full-time employment with AstraZeneca during the conduct of the study and AstraZeneca stock ownership. EAH reports full-time employment with AstraZeneca and AstraZeneca stock ownership. JSB reports full-time employment with AstraZeneca at the time of study and AstraZeneca stock ownership.
Figures

References
-
- Popova T, Manié E, Rieunier G, Caux-Moncoutier V, Tirapo C, Dubois T, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72(21):5454–62. - PubMed
-
- O’Connor MJ. Targeting the DNA damage response in cancer. Mol Cell. 2015;60(4):547–60. - PubMed
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

