Impact of foliar application of phyllosphere yeast strains combined with soil fertilizer application on rice growth and yield
- PMID: 39695904
- PMCID: PMC11657868
- DOI: 10.1186/s40793-024-00635-9
Impact of foliar application of phyllosphere yeast strains combined with soil fertilizer application on rice growth and yield
Abstract
Background: The application of beneficial microbes in agriculture is gaining increasing attention as a means to reduce reliance on chemical fertilizers. This approach can potentially mitigate negative impacts on soil, animal, and human health, as well as decrease climate-changing factors. Among these microbes, yeast has been the least explored, particularly within the phyllosphere compartment. This study addresses this knowledge gap by investigating the potential of phyllosphere yeast to improve rice yield while reducing fertilizer dosage.
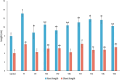
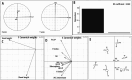
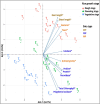
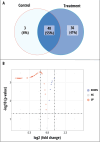
Results: From fifty-two rice yeast phyllosphere isolates, we identified three yeast strains-Rhodotorula paludigena Y1, Pseudozyma sp. Y71, and Cryptococcus sp. Y72-that could thrive at 36 °C and possessed significant multifarious plant growth-promoting traits, enhancing rice root and shoot length upon seed inoculation. These three strains demonstrated favorable compatibility, leading to the creation of a yeast consortium. We assessed the combined effect of foliar application of this yeast consortium and individual strains with two distinct recommended doses of chemical fertilizers (RDCFs) (75 and 100%), as well as RDCFs alone (75 and 100%), in rice maintained in pot-culture and field experiments. The pot-culture experiment investigated the leaf microbial community, plant biochemicals, root and shoot length during the stem elongation, flowering, and dough phases, and yield-related parameters at harvest. The field experiment determined the actual yield. Integrated results from both experiments revealed that the yeast consortium with 75% RDCFs was more effective than the yeast consortium with 100% RDCFs, single strain applications with RDCFs (75 and 100%), and RDCFs alone (75 and 100%). Additionally, this treatment improved leaf metabolite levels compared to control rice plants.
Conclusions: Overall, a 25% reduction in soil chemical fertilizers combined with yeast consortium foliar application improved rice growth, biochemicals, and yield. This study also advances the field of phyllosphere yeast research in agriculture.
Keywords: Crop improvement; Phyllosphere; Plant growth-promoting microbes; Rice; Yeast.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: This material is the authors' own original work, which has not been previously published elsewhere. The paper reflects the authors' own research and analysis in a truthful and complete manner. Competing interests: The authors declare no competing interests.
Figures


References
-
- IPAD. USDA. India Rice Area, Yield and Production. 2024. Retrive from https://ipad.fas.usda.gov/countrysummary/Default.aspx?id=IN&crop=Rice.
-
- FAO. The state of food and agriculture. In: Moving forward on food loss and waste reduction. Rome: FAO; 2019.
-
- Deutsch CA, Tewksbury JJ, Tigchelaar M, Battisti DS, Merrill SC, Huey RB, Naylor RL. Increase in crop losses to insect pests in a warming climate. Science. 2018;361(6405):916–9. 10.1126/science.aat3466. - PubMed
-
- Koli P, Bhardwaj NR, Mahawer SK. Agrochemicals: harmful and beneficial effects of climate changing scenarios. In: Climate change and agricultural ecosystems. New Delhi: Woodhead Publishing; 2019. p. 65–94.
-
- Malhi GS, Kaur M, Kaushik P. Impact of climate change on agriculture and its mitigation strategies: a review. Sustainability. 2021;13(3):1318. 10.3390/su13031318.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
