Molecular characterization and safety properties of multi drug-resistant Escherichia coli O157:H7 bacteriophages
- PMID: 39695941
- PMCID: PMC11657129
- DOI: 10.1186/s12866-024-03691-w
Molecular characterization and safety properties of multi drug-resistant Escherichia coli O157:H7 bacteriophages
Abstract
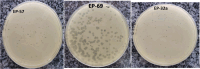
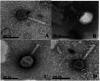
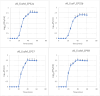
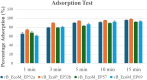
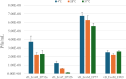
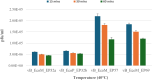
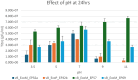
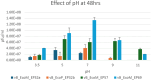
The increase in multi drug resistance (MDR) amongst food-borne pathogens such as Escherichia coli O157:H7, coupled with the upsurge of food-borne infections caused by these pathogens is a major public health concern. Lytic phages have been employed as an alternative to antibiotics for use against food-borne pathogens. However, for effective application, phages should be selectively toxic. Therefore, the objective of this study was to characterise lytic E. coli O157:H7 phages isolated from wastewater as possible biocontrol agents and access their genomes for the absence of genes that denotes virulence, resistance, toxins, and lysogeny using whole genome sequencing. E. coli O157:H7 bacteriophages showed clear plaques ranging in size from 1.0 mm to 2.0 mm. Spot test and Efficiency of plating (EOP) analysis demonstrated that isolated phages could infect various environmental E. coli strains. Four phages; vB_EcoM_EP32a, vB_EcoP_EP32b, vB_EcoM_EP57, and vB_EcoM_EP69 demonstrated broad lytic spectra against E. coli O157:H7 strains. Transmission Electron Microscopy (TEM) showed that all phages have tails and were classified as Caudoviricetes. Growth parameters showed an average latent period of 15 ± 3.8 min, with a maximum burst size of 392 PFU/cell. The phages were stable at three distinct temperatures (4 °C, 28 °C, and 37 °C) and at pH values of 3.5, 5.0, 7.0, 9.0, and 11.0. Based on their morphological distinctiveness, three phages were included in the Whole Genome Sequencing (WGS) analysis. WGS results revealed that E. coli O157:H7 phages (vB_EcoM_EP32a, vB_EcoP_EP32b, and vB_EcoM_EP57) were composed of linear double-stranded DNA (dsDNA) with genome sizes 163,906, 156,698, and 130,723 bp and GC contents of 37.61, 37, and 39% respectively. Phages vB_EcoM_EP32a and vB_EcoP_EP32b genomes were classified under the class Caudoviricetes, Straboviridae family, and the new genus "Phapecoctavirus", while vB_EcoM_EP57 was classified under the class Caudoviricetes, Autographiviridae family. Genome analysis revealed no lysogenic (integrase), virulence, or antimicrobial resistance sequences in all three Escherichia phage genomes. The overall results provided evidence that lytic E. coli O157:H7 bacteriophages in this study, are relatively stable, can infect diverse E. coli strains, and does not contain genes responsible for virulence, resistance, toxins, and lysogeny. Thus, they can be considered as biocontrol candidates against MDR pathogenic E. coli O157:H7 strains in the food industry.
Keywords: E. coli O157:H7; Antibiotic resistance; Biocontrol; Food pathogen; Phage genome; Whole genome sequencing.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: Ethics approval for the study was obtained from the North-West University Animal Research Ethics Committee (NWU-AnimCareREC) and ethics number NWU-00772-23-A5 was assigned to the study. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- Ngene AC, Aguiyi JC, Uzal U, Egbere J, Onyimba IA, Umera AE, et al. Bacteriophages as Bio-control agent against Food-Borne Pathogen E. Coli O157: H7. IOSR J Pharm Biol Sci. 2020;15(2):23–36.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

