Revealing genes related teat number traits via genetic variation in Yorkshire pigs based on whole-genome sequencing
- PMID: 39695943
- PMCID: PMC11657392
- DOI: 10.1186/s12864-024-11109-0
Revealing genes related teat number traits via genetic variation in Yorkshire pigs based on whole-genome sequencing
Abstract
Background: Teat number is one of the most important indicators to evaluate the lactation performance of sows, and increasing the teat number has become an important method to improve the economic efficiency of farms. Therefore, it is particularly important to deeply analyze the genetic mechanism of teat number traits in pigs. In this study, we detected Single Nucleotide Ploymorphism (SNP), Insertion-Deletion (InDel) and Structural variant (SV) by high-coverage whole-genome resequencing data, and selected teat number at birth and functional teat number as two types of teat number traits for genome-wide association study (GWAS) to reveal candidate genes associated with pig teat number traits.
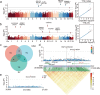
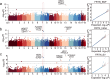
Results: In this study, we used whole genome resequencing data from 560 Yorkshire sows to detect SNPs, InDels and SVs, and performed GWAS for the traits of born teat number and functional teat number, and detected a total of 85 significant variants and screened 214 candidate genes, including HEG1, XYLT1, SULF1, MUC13, VRTN, RAP1A and NPVF. Among them, HEG1 and XYLT1 were the new candidate genes in this study. The co-screening and population validation of multiple traits suggested that HEG1 may have a critical effect on the born teat number.
Conclusion: Our study shows that more candidate genes associated with pig teat number traits can be identified by GWAS with different variant types. Through large population validation, we found that HEG1 may be a new key candidate gene affecting pig teat number traits. In conclusion, the results of this study provide new information for exploring the genetic mechanisms affecting pig teat number traits and genetic improvement of pigs.
Keywords: GWAS; Structural variation; Teat number; Whole genome sequencing.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the Institutional Animal Care and Use Committee of the Northwest A & F University (approval number: NWAFU-314021167). All experiments strictly followed the guidelines of this committee. The experimental animals were not anesthetized or euthanized in this study. All experimental protocols were approved by the aforementioned committee and in accordance with ARRIVE guidelines ( https://arriveguidelines.org ). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Grants and funding
- S2024-YF-YBNY-0752/Key Research and Development Projects of Shaanxi Province
- S2024-YF-YBNY-0752/Key Research and Development Projects of Shaanxi Province
- S2024-YF-YBNY-0752/Key Research and Development Projects of Shaanxi Province
- 2022GD-TSLD-46/Shaanxi Livestock and Poultry Breeding Double-chain Fusion Key Project
- 2022GD-TSLD-46/Shaanxi Livestock and Poultry Breeding Double-chain Fusion Key Project
LinkOut - more resources
Full Text Sources

