NERVE 2.0: boosting the new enhanced reverse vaccinology environment via artificial intelligence and a user-friendly web interface
- PMID: 39695945
- PMCID: PMC11654298
- DOI: 10.1186/s12859-024-06004-0
NERVE 2.0: boosting the new enhanced reverse vaccinology environment via artificial intelligence and a user-friendly web interface
Abstract
Background: Vaccines development in this millennium started by the milestone work on Neisseria meningitidis B, reporting the invention of Reverse Vaccinology (RV), which allows to identify vaccine candidates (VCs) by screening bacterial pathogens genome or proteome through computational analyses. When NERVE (New Enhanced RV Environment), the first RV software integrating tools to perform the selection of VCs, was released, it prompted further development in the field. However, the problem-solving potential of most, if not all, RV programs is still largely unexploited by experimental vaccinologists that impaired by somehow difficult interfaces, requiring bioinformatic skills.
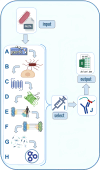
Results: We report here on the development and release of NERVE 2.0 (available at: https://nerve-bio.org ) which keeps the original integrative and modular approach of NERVE, while showing higher predictive performance than its previous version and other web-RV programs (Vaxign and Vaxijen). We renewed some of its modules and added innovative ones, such as Loop-Razor, to recover fragments of promising vaccine candidates or Epitope Prediction for the epitope prediction binding affinities and population coverage. Along with two newly built AI (Artificial Intelligence)-based models: ESPAAN and Virulent. To improve user-friendliness, NERVE was shifted to a tutored, web-based interface, with a noSQL-database to consent the user to submit, obtain and retrieve analysis results at any moment.
Conclusions: With its redesigned and updated environment, NERVE 2.0 allows customisable and refinable bacterial protein vaccine analyses to all different kinds of users.
Keywords: Artificial intelligence; Machine learning; Modular software; Reverse vaccinology; User-friendly website; Vaccine candidates.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Rappuoli R. Reverse vaccinology. Curr Opin Microbiol. 2000;3(5):445–50. - PubMed
-
- Pizza M, Scarlato V, Masignani V, Giuliani MM, Aricò B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, Galeotti CL, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood DW, Jeffries AC, Saunders NJ, Granoff DM, Venter JC, Moxon ER, Grandi G, Rappuoli R. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287(5459):1816–20. - PubMed
-
- Cuypers B, Rappuoli R, Brozzi A. A lean reverse vaccinology pipeline with publicly available bioinformatic tools. Methods Mol Biol. 2023;2673:341–56. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

