DeepMiRBP: a hybrid model for predicting microRNA-protein interactions based on transfer learning and cosine similarity
- PMID: 39695955
- PMCID: PMC11656930
- DOI: 10.1186/s12859-024-05985-2
DeepMiRBP: a hybrid model for predicting microRNA-protein interactions based on transfer learning and cosine similarity
Abstract
Background: Interactions between microRNAs and RNA-binding proteins are crucial for microRNA-mediated gene regulation and sorting. Despite their significance, the molecular mechanisms governing these interactions remain underexplored, apart from sequence motifs identified on microRNAs. To date, only a limited number of microRNA-binding proteins have been confirmed, typically through labor-intensive experimental procedures. Advanced bioinformatics tools are urgently needed to facilitate this research.
Methods: We present DeepMiRBP, a novel hybrid deep learning model specifically designed to predict microRNA-binding proteins by modeling molecular interactions. This innovation approach is the first to target the direct interactions between small RNAs and proteins. DeepMiRBP consists of two main components. The first component employs bidirectional long short-term memory (Bi-LSTM) neural networks to capture sequential dependencies and context within RNA sequences, attention mechanisms to enhance the model's focus on the most relevant features and transfer learning to apply knowledge gained from a large dataset of RNA-protein binding sites to the specific task of predicting microRNA-protein interactions. Cosine similarity is applied to assess RNA similarities. The second component utilizes Convolutional Neural Networks (CNNs) to process the spatial data inherent in protein structures based on Position-Specific Scoring Matrices (PSSM) and contact maps to generate detailed and accurate representations of potential microRNA-binding sites and assess protein similarities.
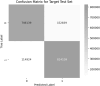
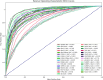
Results: DeepMiRBP achieved a prediction accuracy of 87.4% during training and 85.4% using testing, with an F score of 0.860. Additionally, we validated our method using three case studies, focusing on microRNAs such as miR-451, -19b, -23a, -21, -223, and -let-7d. DeepMiRBP successfully predicted known miRNA interactions with recently discovered RNA-binding proteins, including AGO, YBX1, and FXR2, identified in various exosomes.
Conclusions: Our proposed DeepMiRBP strategy represents the first of its kind designed for microRNA-protein interaction prediction. Its promising performance underscores the model's potential to uncover novel interactions critical for small RNA sorting and packaging, as well as to infer new RNA transporter proteins. The methodologies and insights from DeepMiRBP offer a scalable template for future small RNA research, from mechanistic discovery to modeling disease-related cell-to-cell communication, emphasizing its adaptability and potential for developing novel small RNA-centric therapeutic interventions and personalized medicine.
Keywords: Deep learning; Interaction prediction; MicroRNAs; RNA binding proteins; RNA sorting.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures


References
-
- Azizian S. A data-driven discovery system for studying extracellular micro rna sorting and rna-protein interactions. PhD thesis, The University of Nebraska-Lincoln. 2024.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

