Metagenomic analysis reveals the community composition of the microbiome in different segments of the digestive tract in donkeys and cows: implications for microbiome research
- PMID: 39695983
- PMCID: PMC11656932
- DOI: 10.1186/s12866-024-03696-5
Metagenomic analysis reveals the community composition of the microbiome in different segments of the digestive tract in donkeys and cows: implications for microbiome research
Abstract
Introduction: The intestinal microbiota plays a crucial role in health and disease. This study aimed to assess the composition and functional diversity of the intestinal microbiota in donkeys and cows by examining samples collected from different segments of the digestive tract using two distinct techniques: direct swab sampling and faecal sampling.
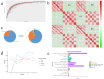
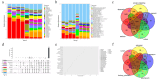
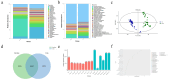
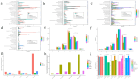
Results: In this study, we investigated and compared the effects of multiple factors on the composition and function of the intestinal microbial community. Approximately 300 GB of metagenomic sequencing data from 91 samples obtained from various segments of the digestive tract were used, including swabs and faecal samples from monogastric animals (donkeys) and polygastric animals (cows). We assembled 4,004,115 contigs for cows and 2,938,653 contigs for donkeys, with a total of 9,060,744 genes. Our analysis revealed that, compared with faecal samples, swab samples presented a greater abundance of Bacteroidetes, whereas faecal samples presented a greater abundance of Firmicutes. Additionally, we observed significant variations in microbial composition among different digestive tract segments in both animals. Our study identified key bacterial species and pathways via different methods and provided evidence that multiple factors can influence the microbial composition. These findings provide new insights for the accurate characterization of the composition and function of the gut microbiota in microbiome research.
Conclusions: The results obtained by both sampling methods in the present study revealed that the composition and function of the intestinal microbiota in donkeys and cows exhibit species-specific and region-specific differences. These findings highlight the importance of using standardized sampling protocols to ensure accurate and consistent characterization of the intestinal microbiota in various animal species. The implications and underlying mechanisms of these associations provide multiple perspectives for future microbiome research.
Keywords: Digestive tract segments; Intestinal microbiota; Metagenomic analysis; Microbiota composition.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Our animal experiments were approved and authorized by the Ethics Committee of the Laboratory of Animal Sciences of Peking Union Medical College (no. XZG22001). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Tee MZ, Er YX, Easton AV, Yap NJ, Lee IL, Devlin J, Chen Z, Ng KS, Subramanian P, Angelova A, Oyesola O, Sargsian S, Ngui R, Beiting DP, Boey CCM, Chua KH, Cadwell K, Lim YAL, Loke P, Lee SC. Gut microbiome of helminth-infected indigenous Malaysians is context dependent. Microbiome. 2022;10:214. 10.1186/s40168-022-01385-x. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

