Exploring autism spectrum disorder and co-occurring trait associations to elucidate multivariate genetic mechanisms and insights
- PMID: 39696186
- PMCID: PMC11658126
- DOI: 10.1186/s12888-024-06392-w
Exploring autism spectrum disorder and co-occurring trait associations to elucidate multivariate genetic mechanisms and insights
Abstract
Background: Autism spectrum disorder (ASD) is a partially heritable neurodevelopmental trait, and people with ASD may also have other co-occurring trait such as ADHD, anxiety disorders, depression, mental health issues, learning difficulty, physical health traits and communication challenges. The concomitant development of ASD and other neurological traits is assumed to result from a complex interplay between genetics and the environment. However, only a limited number of studies have performed multivariate genome-wide association studies (GWAS) for ASD.
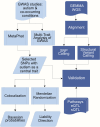
Methods: We conducted to-date the largest multivariate GWAS on ASD and 8 ASD co-occurring traits (ADHD, ADHD childhood, anxiety stress (ASDR), bipolar (BIP), disruptive behaviour (DBD), educational attainment (EA), major depression, and schizophrenia (SCZ)) using summary statistics from leading studies. Multivariate associations and central traits were further identified. Subsequently, colocalization and Mendelian randomization (MR) analysis were performed on the associations identified with the central traits containing ASD. To further validate our findings, pathway and quantified trait loci (QTL) resources as well as independent datasets consisting of 112 (45 probands) whole genome sequence data from the GEMMA project were utilized.
Results: Multivariate GWAS resulted in 637 significant associations (p < 5e-8), among which 322 are reported for the first time for any trait. 37 SNPs were identified to contain ASD and one or more traits in their central trait set, including variants mapped to known SFARI ASD genes MAPT, CADPS and NEGR1 as well as novel ASD genes KANSL1, NSF and NTM, associated with immune response, synaptic transmission, and neurite growth respectively. Mendelian randomization analyses found that genetic liability for ADHD childhood, ASRD and DBT has causal effects on the risk of ASD while genetic liability for ASD has causal effects on the risk of ADHD, ADHD childhood, BIP, WA, MDD and SCZ. Frequency differences of SNPs found in NTM and CADPS genes, respectively associated with neurite growth and neural/endocrine calcium regulation, were found between GEMMA ASD probands and controls. Pathway, QTL and cell type enrichment implicated microbiome, enteric inflammation, and central nervous system enrichments.
Conclusions: Our study, combining multivariate GWAS with systematic decomposition, identified novel genetic associations related to ASD and ASD co-occurring driver traits. Statistical tests were applied to discern evidence for shared and interpretable liability between ASD and co-occurring traits. These findings expand upon the current understanding of the complex genetics regulating ASD and reveal insights of neuronal brain disruptions potentially driving development and manifestation.
Keywords: ASD; ASD genetically correlated traits; GEMMA; Mendelian randomization; Multivariate GWAS.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The GEMMA study was approved by the relevant ethics committee of each enrolling country. Particularly, CE Campania Sud (IRB n.30/2019) for Italy; Partners Human Research (IRB ver.01/04/2019) for USA; and Clinical Research Ethics Committee of Galway University Hospital (IRB n. C.A. 2127/19) for Ireland. A written consent form will be signed by each participant or their legal representative. Consent for publication: Consent, relevant to GEMMA subjects, is granted and signed by each participant or their legal representative. Competing interests: The authors declare no competing interests.
Figures

References
-
- Ashburner J, Ziviani J, and Sylvia Rodger. Surviving in the mainstream: capacity of children with Autism Spectrum disorders to perform academically and regulate their emotions and behavior at School. Res Autism Spectr Disorders. 2010;4(1):18–27.
-
- Baio J, Wiggins L, Christensen DL, Maenner MJ, Daniels J, Warren Z, Kurzius-Spencer M, et al. Prevalence of Autism Spectrum Disorder among children aged 8 years - Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2014. Morbidity Mortal Wkly Rep Surveillance Summaries. 2018;67(6):1–23. - PMC - PubMed
-
- Boorstein HC. Gastrointestinal and feeding problems in Young children: a comparison between Autism Spectrum disorders and other Developmental delays. University of Connecticut; 2008.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

