ZO-1 boosts the in vitro self-renewal of pre-haematopoietic stem cells from OCT4-reprogrammed human hair follicle mesenchymal stem cells through cytoskeleton remodeling
- PMID: 39696518
- PMCID: PMC11658245
- DOI: 10.1186/s13287-024-04080-w
ZO-1 boosts the in vitro self-renewal of pre-haematopoietic stem cells from OCT4-reprogrammed human hair follicle mesenchymal stem cells through cytoskeleton remodeling
Erratum in
-
Correction: ZO-1 boosts the in vitro self-renewal of pre-haematopoietic stem cells from OCT4-reprogrammed human hair follicle mesenchymal stem cells through cytoskeleton remodeling.Stem Cell Res Ther. 2025 Mar 8;16(1):126. doi: 10.1186/s13287-025-04259-9. Stem Cell Res Ther. 2025. PMID: 40055776 Free PMC article. No abstract available.
Abstract
Background: The challenge of expanding haematopoietic stem/progenitor cells (HSPCs) in vitro has limited their clinical application. Human hair follicle mesenchymal stem cells (hHFMSCs) can be reprogrammed to generate intermediate stem cells by transducing OCT4 (hHFMSCsOCT4) and pre-inducing with FLT3LG/SCF, and differentiated into erythrocytes. These intermediate cells exhibit gene expression patterns similar to pre-HSCs, making them promising for artificial haematopoiesis. However, further investigation is required to elucidate the in vitro proliferation ability and mechanism underlying the self-renewal of pre-HSCs derived from hHFMSCs.
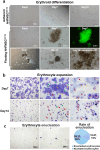
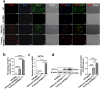
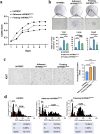
Methods: hHFMSCsOCT4 were pre-treated with FLT3LG and SCF cytokines, followed by characterization and isolation of the floating cell subsets for erythroid differentiation through stimulation with hematopoietic cytokines and nutritional factors. Cell adhesion was assessed through disassociation and adhesion assays. OCT4 expression levels were measured using immunofluorescence staining, RT-qPCR, and Western blotting. RNA sequencing and Gene Ontology (GO) enrichment analysis were then conducted to identify proliferation-related biological processes. Proliferative capacity was evaluated through CCK-8, colony formation assays, Ki67 index, and cell cycle analysis. Cytoskeleton was observed through Wright‒Giemsa, Coomassie brilliant blue, and phalloidin staining. Expression of adherens junction (AJ) core members was confirmed through RT‒qPCR, Western blotting, and immunofluorescence staining before and after ZO-1 knockdown. A regulatory network was constructed to determine relationships among cytoskeleton, proliferation, and the AJ pathway. Student's t tests (GraphPad Prism 8.0.2) were used for group comparisons. The results were considered significant at P < 0.05.
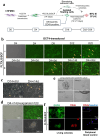
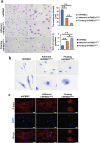
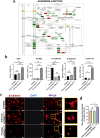
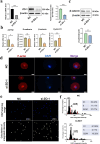
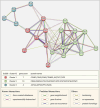
Results: Pre-treatment of hHFMSCsOCT4 with FLT3LG and SCF leads to the emergence of floating cell subsets exhibiting small, globoid morphology, suspended above adherent cells, forming colonies, and displaying minimal expression of CD45. Excessive OCT4 expression weakens adhesion in floating hHFMSCsOCT4. Floating cells moderately enhanced proliferation and undergo cytoskeleton remodelling, with increased contraction and aggregation of F-actin near the nucleus. The upregulation of ZO-1 could impact the expressions of F-actin, E-cadherin, and β-catenin genes, as well as the nuclear positioning of β-catenin, leading to variations in the cytoskeleton and cell cycle. Finally, a regulatory network revealed that the AJ pathway cored with ZO-1 critically bridges cytoskeletal remodelling and haematopoiesis-related proliferation in a β-catenin-dependent manner.
Conclusions: ZO-1 improved the self-renewal of pre-HSCs from OCT4-overexpressing hHFMSCs by remodeling the cytoskeleton via the ZO-1-regulated AJ pathway, suggesting floating hHFMSCsOCT4 as the promising seed cells for artificial hematopoiesis.
Keywords: Adherens junction; Human hair follicle mesenchymal stem cells; Pre-haematopoietic stem cells; Reprogramming; Self-renewal.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Informed consent was obtained from the donors to isolated hHFMSCs for research use. All experiments were approved by the Medical Ethics Committee of Qingdao Municipal Hospital. Title of the approved project: OCT4 facilitates the transdifferentiation of human hair follicle stem cells into erythrocytes by remodeling the cytoskeleton. Name of the institutional approval committee or unit: Medical Ethics Committee/IRB of Qingdao Municipal Hospital. Approval number: 2024-KY-077, Date of approval: Oct 16, 2024. Consent for publication: Informed consent was obtained from the donors to isolated hHFMSCs for research use. All authors have reviewed the manuscript and approved the publication. Competing interests: The authors declare that they have no competing interests.
Figures
Similar articles
-
RNA-seq reveals tight junction-relevant erythropoietic fate induced by OCT4 in human hair follicle mesenchymal stem cells.Stem Cell Res Ther. 2020 Oct 27;11(1):454. doi: 10.1186/s13287-020-01976-1. Stem Cell Res Ther. 2020. PMID: 33109258 Free PMC article.
-
OCT4 maintains self-renewal and reverses senescence in human hair follicle mesenchymal stem cells through the downregulation of p21 by DNA methyltransferases.Stem Cell Res Ther. 2019 Jan 15;10(1):28. doi: 10.1186/s13287-018-1120-x. Stem Cell Res Ther. 2019. PMID: 30646941 Free PMC article.
-
PBX homeobox 1 enhances hair follicle mesenchymal stem cell proliferation and reprogramming through activation of the AKT/glycogen synthase kinase signaling pathway and suppression of apoptosis.Stem Cell Res Ther. 2019 Aug 23;10(1):268. doi: 10.1186/s13287-019-1382-y. Stem Cell Res Ther. 2019. PMID: 31443676 Free PMC article.
-
N-cadherin-based adherens junction regulates the maintenance, proliferation, and differentiation of neural progenitor cells during development.Cell Adh Migr. 2015;9(3):183-92. doi: 10.1080/19336918.2015.1005466. Epub 2015 Apr 14. Cell Adh Migr. 2015. PMID: 25869655 Free PMC article. Review.
-
The tortoise and the hair: slow-cycling cells in the stem cell race.Cell. 2009 May 29;137(5):811-9. doi: 10.1016/j.cell.2009.05.002. Cell. 2009. PMID: 19490891 Free PMC article. Review.
Cited by
-
Correction: ZO-1 boosts the in vitro self-renewal of pre-haematopoietic stem cells from OCT4-reprogrammed human hair follicle mesenchymal stem cells through cytoskeleton remodeling.Stem Cell Res Ther. 2025 Mar 8;16(1):126. doi: 10.1186/s13287-025-04259-9. Stem Cell Res Ther. 2025. PMID: 40055776 Free PMC article. No abstract available.
References
-
- C D. Blood stem cells produced in vast quantities in the lab. Nature. 2019;570(7759):17–8. - PubMed
-
- Sakurai MIK, Ito R, Wilkinson AC, Kimura T, Mizutani E, Nishikii H, Sudo K, Becker HJ, Takemoto H, Sano T, Kataoka K, Takahashi S, Nakamura Y, Kent DG, Iwama A, Chiba S, Okamoto S, Nakauchi H, Yamazaki S. Chemically defined cytokine-free expansion of human haematopoietic stem cells. Nature. 2023;615(7950):127–33. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

