Diverse microbiome functions, limited temporal variation and substantial genomic conservation within sedimentary and granite rock deep underground research laboratories
- PMID: 39696556
- PMCID: PMC11657941
- DOI: 10.1186/s40793-024-00649-3
Diverse microbiome functions, limited temporal variation and substantial genomic conservation within sedimentary and granite rock deep underground research laboratories
Abstract
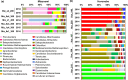
Background: Underground research laboratories (URLs) provide a window on the deep biosphere and enable investigation of potential microbial impacts on nuclear waste, CO2 and H2 stored in the subsurface. We carried out the first multi-year study of groundwater microbiomes sampled from defined intervals between 140 and 400 m below the surface of the Horonobe and Mizunami URLs, Japan.
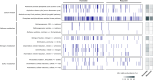
Results: We reconstructed draft genomes for > 90% of all organisms detected over a four year period. The Horonobe and Mizunami microbiomes are dissimilar, likely because the Mizunami URL is hosted in granitic rock and the Horonobe URL in sedimentary rock. Despite this, hydrogen metabolism, rubisco-based CO2 fixation, reduction of nitrogen compounds and sulfate reduction are well represented functions in microbiomes from both URLs, although methane metabolism is more prevalent at the organic- and CO2-rich Horonobe URL. High fluid flow zones and proximity to subsurface tunnels select for candidate phyla radiation bacteria in the Mizunami URL. We detected near-identical genotypes for approximately one third of all genomically defined organisms at multiple depths within the Horonobe URL. This cannot be explained by inactivity, as in situ growth was detected for some bacteria, albeit at slow rates. Given the current low hydraulic conductivity and groundwater compositional heterogeneity, ongoing inter-site strain dispersal seems unlikely. Alternatively, the Horonobe URL microbiome homogeneity may be explained by higher groundwater mobility during the last glacial period. Genotypically-defined species closely related to those detected in the URLs were identified in three other subsurface environments in the USA. Thus, dispersal rates between widely separated underground sites may be fast enough relative to mutation rates to have precluded substantial divergence in species composition. Species overlaps between subsurface locations on different continents constrain expectations regarding the scale of global subsurface biodiversity.
Conclusions: Our analyses reveal microbiome stability in the sedimentary rocks and surprising microbial community compositional and genotypic overlap over sites separated by hundreds of meters of rock, potentially explained by dispersal via slow groundwater flow or during a prior hydrological regime. Overall, microbiome and geochemical stability over the study period has important implications for underground storage applications.
Keywords: Granite; Groundwater; Metagenomics microbiome; Sedimentary rocks; Stability; Underground research laboratory.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
Deep microbial life in high-quality granitic groundwater from geochemically and geographically distinct underground boreholes.Environ Microbiol Rep. 2016 Apr;8(2):285-94. doi: 10.1111/1758-2229.12379. Epub 2016 Jan 28. Environ Microbiol Rep. 2016. PMID: 26743638
-
Deep groundwater physicochemical components affecting actinide migration.Chemosphere. 2022 Feb;289:133181. doi: 10.1016/j.chemosphere.2021.133181. Epub 2021 Dec 4. Chemosphere. 2022. PMID: 34875295
-
Spatial and Temporal Constraints on the Composition of Microbial Communities in Subsurface Boreholes of the Edgar Experimental Mine.Microbiol Spectr. 2021 Dec 22;9(3):e0063121. doi: 10.1128/Spectrum.00631-21. Epub 2021 Nov 10. Microbiol Spectr. 2021. PMID: 34756066 Free PMC article.
-
Paleo-Rock-Hosted Life on Earth and the Search on Mars: A Review and Strategy for Exploration.Astrobiology. 2019 Oct;19(10):1230-1262. doi: 10.1089/ast.2018.1960. Epub 2019 Jun 25. Astrobiology. 2019. PMID: 31237436 Free PMC article. Review.
-
Artificial subsurface lithoautotrophic microbial ecosystems and gas storage in deep subsurface.FEMS Microbiol Ecol. 2024 Oct 25;100(11):fiae142. doi: 10.1093/femsec/fiae142. FEMS Microbiol Ecol. 2024. PMID: 39448371 Free PMC article. Review.
Cited by
-
Sequence Modeling Is Not Evolutionary Reasoning.bioRxiv [Preprint]. 2025 Jun 27:2025.01.17.633626. doi: 10.1101/2025.01.17.633626. bioRxiv. 2025. PMID: 39896621 Free PMC article. Preprint.
References
-
- Pedersen K. Subterranean microorganisms and radioactive waste disposal in Sweden. Eng Geol. 1999;52:163–76.
-
- Stroes-Gascoyne S, Hamon C, Maak P, Russell S. The effects of the physical properties of highly compacted smectitic clay (bentonite) on the culturability of indigenous microorganisms. Appl Clay Sci. 2010;47:155–62.
Grants and funding
- JPJ007597/The Ministry of Economy, Trade and Industry of Japan
- 19K05342/the Japan Society for the Promotion of Science
- OCE2049478/National Science Foundation
- DE-AC02-05CH11231/the Watershed Function Scientific Focus Area funded by the U.S. Department of Energy
- 2230766/NSF "Four Networks for Geologic Hydrogen Storage"
LinkOut - more resources
Full Text Sources
