Transcriptome-aligned metabolic profiling by SERSome reflects biological changes following mesenchymal stem cells expansion
- PMID: 39696645
- PMCID: PMC11656856
- DOI: 10.1186/s13287-024-04109-0
Transcriptome-aligned metabolic profiling by SERSome reflects biological changes following mesenchymal stem cells expansion
Abstract
Background: Mesenchymal stem cells (MSCs) are widely applied in the treatment of various clinical diseases and in the field of medical aesthetics. However, MSCs exhibit greater heterogeneity limited stability, and more complex molecular and mechanistic characteristics compared to conventional drugs, making rapid and precise monitoring more challenging.
Methods: Surface-enhanced Raman spectroscopy (SERS) is an ultrasensitive, tractable and low-cost fingerprinting technique capable of identifying a wide range of molecules related to biological processes. Here, we employed SERS for reproducible quantification of ultralow concentrations of molecules and utilized spectral sets, termed SERSomes, for robust and comprehensive intracellular multi-metabolite profiling.
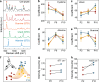
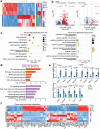
Results: We revealed that with increasing passage number, there is a gradual decline in cell expansion efficiency, accompanied by significant changes in intracellular amino acids, purines, and pyrimidines. By integrating these metabolic features detected by SERS with transcriptomic data, we established a correlation between SERS signals and biological changes, as well as differentially expressed genes.
Conclusion: In this study, we explore the application of SERS technique to provide robust metabolic characteristics of MSCs across different passages and donors. These results demonstrate the effectiveness of SERSome in reflecting biological characteristics. Due to its sensitivity, adaptability, low cost, and feasibility for miniaturized instrumentation throughout pretreatment, measurement, and analysis, the label-free SERSome technique is suitable for monitoring MSC expansion and offers significant advantages for large-scale MSC manufacturing.
Keywords: Mesenchymal stem cells (MSCs); Metabolism; Surface-enhanced Raman spectroscopy (SERS); Transcriptome.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was conducted at the School of Biomedical Engineering and Clinical Stem Cell Research Center, Shanghai Jiao Tong University. Approval was granted for a project titled “Cancer Immunotherapy Using Umbilical Cord Mesenchymal Stem Cells” in 2021 by the Renji Hospital Ethics Committee (No. KY2021-027). All participants provided informed consent. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Grants and funding
- 82203255/National Natural Science Foundation of China
- 82272054/National Natural Science Foundation of China
- W2431055/National Natural Science Foundation of China
- ZJ2021-ZD-007/Major Projects of Special Development Fund in Shanghai Zhangjiang National Independent Innovation Demonstration Zone
- YG2024LC09/Shanghai Key Laboratory of Gynecologic Oncology, Shanghai Jiao Tong University
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

