Structural variant allelic heterogeneity in MECP2 duplication syndrome provides insight into clinical severity and variability of disease expression
- PMID: 39696717
- PMCID: PMC11658439
- DOI: 10.1186/s13073-024-01411-7
Structural variant allelic heterogeneity in MECP2 duplication syndrome provides insight into clinical severity and variability of disease expression
Abstract
Background: MECP2 Duplication Syndrome, also known as X-linked intellectual developmental disorder Lubs type (MRXSL; MIM: 300260), is a neurodevelopmental disorder caused by copy number gains spanning MECP2. Despite varying genomic rearrangement structures, including duplications and triplications, and a wide range of duplication sizes, no clear correlation exists between DNA rearrangement and clinical features. We had previously demonstrated that up to 38% of MRXSL families are characterized by complex genomic rearrangements (CGRs) of intermediate complexity (2 ≤ copy number variant breakpoints < 5), yet the impact of these genomic structures on regulation of gene expression and phenotypic manifestations have not been investigated.
Methods: To study the role of the genomic rearrangement structures on an individual's clinical phenotypic variability, we employed a comprehensive genomics, transcriptomics, and deep phenotyping analysis approach on 137 individuals affected by MRXSL. Genomic structural information was correlated with transcriptomic and quantitative phenotypic analysis using Human Phenotype Ontology (HPO) semantic similarity scores.
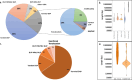
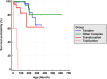
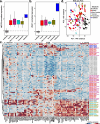
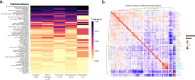
Results: Duplication sizes in the cohort ranging from 64.6 kb to 16.5 Mb were classified into four categories comprising of tandem duplications (48%), terminal duplications (22%), inverted triplications (20%), and other CGRs (10%). Most of the terminal duplication structures consist of translocations (65%) followed by recombinant chromosomes (23%). Notably, 65% of de novo events occurred in the Terminal duplication group in contrast with 17% observed in Tandem duplications. RNA-seq data from lymphoblastoid cell lines indicated that the MECP2 transcript quantity in MECP2 triplications is statistically different from all duplications, but not between other classes of genomic structures. We also observed a significant (p < 0.05) correlation (Pearson R = 0.6, Spearman p = 0.63) between the log-transformed MECP2 RNA levels and MECP2 protein levels, demonstrating that genomic aberrations spanning MECP2 lead to altered MECP2 RNA and MECP2 protein levels. Genotype-phenotype analyses indicated a gradual worsening of phenotypic features, including overall survival, developmental levels, microcephaly, epilepsy, and genitourinary/eye abnormalities in the following order: Tandem duplications, Other complex duplications, Terminal duplications/Translocations, and Triplications encompassing MECP2.
Conclusion: In aggregate, this combined analysis uncovers an interplay between MECP2 dosage, genomic rearrangement structure and phenotypic traits. Whereas the level of MECP2 is a key determinant of the phenotype, the DNA rearrangement structure can contribute to clinical severity and disease expression variability. Employing this type of analytical approach will advance our understanding of the impact of genomic rearrangements on genomic disorders and may help guide more targeted therapeutic approaches.
Keywords: MECP2 duplication syndrome; Clinical severity; MRXSL; Survival; Tandem duplication; Terminal duplication.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study is approved by Baylor College of Medicine (BCM) and Pacific Northwest Research Institute (PNRI). For patients who were evaluated at TCH-BBC Rett Center, clinical information was obtained by retrospective chart review using H-46044 protocol approved by Baylor College of Medicine’s (BCM) Institutional Review Board (IRB). We used BCM IRB approved H-32407 and H-47281/PNRI WIRB #20202158 protocols to clinically examine patients on a research basis. For genomic studies, participants were consented according to the IRB at BCM approved protocols: H-29697, H-20268, H-18122, and H-26667 or H-47281/Pacific Northwest Research Institute WIRB #20202158. The research was conducted in compliance with the principles of Helsinki Declaration. Consent for publication: All subjects gave consent for participation into our study and publication of genomic and clinical information. Competing interests: BCM and Miraca Holdings have formed a joint venture with shared ownership and governance of BG, which performs clinical microarray analysis (CMA), clinical ES (cES), and clinical biochemical studies. J.R.L. serves on the Scientific Advisory Board of the BG. J.R.L. has stock ownership in 23andMe, is a paid consultant for Genomics International, and is a coinventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, genomic disorders and bacterial genomic fingerprinting. D.P. and M.B.R. provide consulting service for Ionis Pharmaceuticals. The remaining authors declare that they have no competing interests.
Figures


References
-
- Lagger S, Connelly JC, Schweikert G, Webb S, Selfridge J, Ramsahoye BH, Yu M, He C, Sanguinetti G, Sowers LC, et al. MeCP2 recognizes cytosine methylated tri-nucleotide and di-nucleotide sequences to tune transcription in the mammalian brain. PLoS Genet. 2017;13:e1006793. 10.1371/journal.pgen.1006793. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

