Tubulin-Based Microtentacles Aid in Heterotypic Clustering of Neutrophil-Differentiated HL-60 Cells and Breast Tumor Cells
- PMID: 39696759
- PMCID: PMC11809343
- DOI: 10.1002/advs.202409260
Tubulin-Based Microtentacles Aid in Heterotypic Clustering of Neutrophil-Differentiated HL-60 Cells and Breast Tumor Cells
Abstract
Circulating tumor cells (CTCs) travel through the vasculature to seed secondary sites and serve as direct precursors of metastatic outgrowth for many solid tumors. Heterotypic cell clusters form between CTCs and white blood cells (WBCs) and recent studies report that a majority of these WBCs are neutrophils in patient and mouse models. The lab discovered that CTCs produce tubulin-based protrusions, microtentacles (McTNs), which promote reattachment, retention in distant sites during metastasis and formation of tumor cell clusters. Neutrophil-CTC clusters help CTCs survive the harsh vascular environment to promote successful metastasis, however, the specific mechanism of this interaction is not fully understood. Utilizing TetherChip technology, it is found that primary and differentiated neutrophils produce McTNs composed of detyrosinated and acetylated α-tubulin and vimentin. Neutrophil McTNs aid in cluster formation, migration, and reattachment, which are suppressed with the tubulin-depolymerizing agent, Vinorelbine. Co-culturing differentiated neutrophils and tumor cells formed heterotypic clusters that enhanced migration. CTC-neutrophil clusters have higher metastatic efficiency, and by demonstrating that neutrophils form McTNs, a new possible mechanism for how neutrophils interact with tumor cells is revealed. These findings further support the idea that developing cluster-disrupting therapies can provide a new targeted strategy to reduce the metastatic potential of cancer cells.
Keywords: HL‐60 cells; TetherChip; circulating tumor cells; heterotypic cluster; metastasis; microtentacles; neutrophils.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The University of Maryland School of Medicine owns patents on the subject of microtentacles and microfluidic cell tethering that list one of the authors on the current manuscript (Stuart S. Martin) as an inventor.
Figures



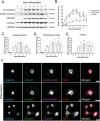


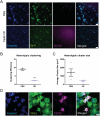

References
MeSH terms
Substances
Grants and funding
- R01-CA154624/NH/NIH HHS/United States
- CH-649-CRF/Maryland Department of Health's Cigarette Restitution Fund Program
- T32 GM008181/GM/NIGMS NIH HHS/United States
- P30-CA134274/Greenebaum Comprehensive Cancer Center
- 5T32GM008181-3334/GM/NIGMS NIH HHS/United States
- P30 CA134274/CA/NCI NIH HHS/United States
- I01 BX002746/BX/BLRD VA/United States
- I01-BX002746/U.S. Department of Veterans Affairs
- 5T32CA154274-13/CA/NCI NIH HHS/United States
- R01 CA154624/CA/NCI NIH HHS/United States
- R01-HL152210/HL/NHLBI NIH HHS/United States
- RSG-18-028-01-CSM/American Cancer Society
- T32 CA154274/CA/NCI NIH HHS/United States
- R01 HL152210/HL/NHLBI NIH HHS/United States
- R01 CA124704/CA/NCI NIH HHS/United States
- R01-CA124704/NH/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
