Advancements in Global Phosphoproteomics Profiling: Overcoming Challenges in Sensitivity and Quantification
- PMID: 39696887
- PMCID: PMC11735659
- DOI: 10.1002/pmic.202400087
Advancements in Global Phosphoproteomics Profiling: Overcoming Challenges in Sensitivity and Quantification
Abstract
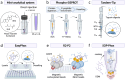
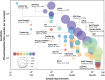
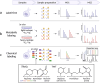
Protein phosphorylation introduces post-genomic diversity to proteins, which plays a crucial role in various cellular activities. Elucidation of system-wide signaling cascades requires high-performance tools for precise identification and quantification of dynamics of site-specific phosphorylation events. Recent advances in phosphoproteomic technologies have enabled the comprehensive mapping of the dynamic phosphoproteomic landscape, which has opened new avenues for exploring cell type-specific functional networks underlying cellular functions and clinical phenotypes. Here, we provide an overview of the basics and challenges of phosphoproteomics, as well as the technological evolution and current state-of-the-art global and quantitative phosphoproteomics methodologies. With a specific focus on highly sensitive platforms, we summarize recent trends and innovations in miniaturized sample preparation strategies for micro-to-nanoscale and single-cell profiling, data-independent acquisition mass spectrometry (DIA-MS) for enhanced coverage, and quantitative phosphoproteomic pipelines for deep mapping of cell and disease biology. Each aspect of phosphoproteomic analysis presents unique challenges and opportunities for improvement and innovation. We specifically highlight evolving phosphoproteomic technologies that enable deep profiling from low-input samples. Finally, we discuss the persistent challenges in phosphoproteomic technologies, including the feasibility of nanoscale and single-cell phosphoproteomics, as well as future outlooks for biomedical applications.
Keywords: data‐independent acquisition; mass spectrometry; phosphoproteomics; protein phosphorylation.
© 2024 The Author(s). Proteomics published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Mapping Nanoscale-To-Single-Cell Phosphoproteomic Landscape by Chip-DIA.Adv Sci (Weinh). 2025 Jan;12(1):e2402421. doi: 10.1002/advs.202402421. Epub 2024 Oct 14. Adv Sci (Weinh). 2025. PMID: 39401432 Free PMC article.
-
Analytical challenges translating mass spectrometry-based phosphoproteomics from discovery to clinical applications.Electrophoresis. 2014 Dec;35(24):3430-40. doi: 10.1002/elps.201400153. Epub 2014 Jul 10. Electrophoresis. 2014. PMID: 24890697 Free PMC article. Review.
-
[Advances in analysis techniques of phosphoproteome].Sheng Wu Gong Cheng Xue Bao. 2003 Mar;19(2):244-8. Sheng Wu Gong Cheng Xue Bao. 2003. PMID: 15966331 Review. Chinese.
-
Plant phosphoproteomics: a long road ahead.Proteomics. 2006 Oct;6(20):5517-28. doi: 10.1002/pmic.200600232. Proteomics. 2006. PMID: 16991200 Review.
-
Recent advances and challenges in plant phosphoproteomics.Proteomics. 2015 Mar;15(5-6):1127-41. doi: 10.1002/pmic.201400410. Epub 2015 Jan 21. Proteomics. 2015. PMID: 25429768 Review.
Cited by
-
Evaluating First-Pass, High Protein Capacity Desalting Techniques For Phosphoproteomics Applications.bioRxiv [Preprint]. 2025 Jun 3:2025.06.03.657744. doi: 10.1101/2025.06.03.657744. bioRxiv. 2025. PMID: 40501759 Free PMC article. Preprint.
-
Integrated Single-Tip IMAC-HILIC Enables Simultaneous Analysis of Plant Phosphoproteomics and N-Glycoproteomics.J Proteome Res. 2025 Jul 4;24(7):3560-3568. doi: 10.1021/acs.jproteome.5c00185. Epub 2025 Jun 23. J Proteome Res. 2025. PMID: 40548449 Free PMC article.
References
-
- Humphrey S. J., James D. E., and Mann M., “Protein Phosphorylation: A Major Switch Mechanism for Metabolic Regulation,” Trends in Endocrinology and Metabolism 26 (2015): 676–687. - PubMed
-
- Needham E. J., Parker B. L., Burykin T., James D. E., and Humphrey S. J., “Illuminating the Dark Phosphoproteome,” Science Signaling (2019): 12. - PubMed
-
- Sharma K., D'souza R. C. J., Tyanova S., et al., “Ultradeep Human Phosphoproteome Reveals a Distinct Regulatory Nature of Tyr and Ser/Thr‐Based Signaling,” Cell Reports 8 (2014): 1583–1594. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

