Machine learning-based diagnostic model of lymphatics-associated genes for new therapeutic target analysis in intervertebral disc degeneration
- PMID: 39697339
- PMCID: PMC11652530
- DOI: 10.3389/fimmu.2024.1441028
Machine learning-based diagnostic model of lymphatics-associated genes for new therapeutic target analysis in intervertebral disc degeneration
Abstract
Background: Low back pain resulting from intervertebral disc degeneration (IVDD) represents a significant global social problem. There are notable differences in the distribution of lymphatic vessels (LV) in normal and pathological intervertebral discs. Nevertheless, the molecular mechanisms of lymphatics-associated genes (LAGs) in the development of IVDD remain unclear. An in-depth exploration of this area will help to reveal the biological and clinical significance of LAGs in IVDD and may lead to the search for new therapeutic targets for IVDD.
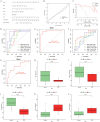
Methods: Data sets were obtained from the Gene Expression Omnibus (GEO) database. Following quality control and normalization, the datasets (GSE153761, GSE147383, and GSE124272) were merged to form the training set, with GSE150408 serving as the validation set. LAGs from GeneCards, MSigDB, Gene Ontology, and KEGG database. The Venn diagram was employed to identify differentially expressed lymphatic-associated genes (DELAGs) that were differentially expressed in the normal and IVDD groups. Subsequently, four machine learning algorithms (SVM-RFE, Random Forest, XGB, and GLM) were used to select the method to construct the diagnostic model. The receiver operating characteristic (ROC) curve, nomogram, and Decision Curve Analysis (DCA) were used to evaluate the model effect. In addition, we constructed a potential drug regulatory network and competitive endogenous RNA (ceRNA) network for key LAGs.
Results: A total of 15 differentially expressed LAGs were identified. By comparing four machine learning methods, the top five genes of importance in the XGB model (MET, HHIP, SPRY1, CSF1, TOX) were identified as lymphatics-associated gene diagnostic signatures. This signature was used to predict the diagnosis of IVDD with strong accuracy and an area under curve (AUC) value of 0.938. Furthermore, the diagnostic model was validated in an external dataset (GSE150408), with an AUC value of 0.772. The nomogram and DCA further prove that the diagnosis model has good performance and predictive value. Additionally, drug regulatory networks and ceRNA networks were constructed, revealing potential therapeutic drugs and post-transcriptional regulatory mechanisms.
Conclusion: We developed and validated a lymphatics-associated genes diagnostic model by machine learning algorithms that effectively identify IVDD patients. These five key LAGs may be potential therapeutic targets for IVDD patients.
Keywords: diagnostic model; intervertebral disc degeneration; lymphatic-associated gene; machine learning; therapeutic target.
Copyright © 2024 Lin, Li, Wang, Zheng, Hu, Zhang, Song and Zhou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

