Geranylgeranyl diphosphate synthase inhibition impairs osteoclast differentiation, morphology, and resorptive activity
- PMID: 39697524
- PMCID: PMC11653010
- DOI: 10.1093/jbmrpl/ziae133
Geranylgeranyl diphosphate synthase inhibition impairs osteoclast differentiation, morphology, and resorptive activity
Abstract
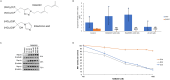
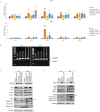
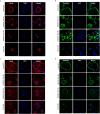
Nitrogen bisphosphonates, such as zoledronic acid, target the enzyme farnesyl diphosphate synthase (FDPS) in the isoprenoid biosynthetic pathway (IBP), and are the frontline treatment for osteolytic bone diseases. A strong affinity of these agents for bone limits their distribution out of the skeleton. Geranylgeranyl diphosphate synthase (GGDPS) is directly downstream to FDPS in the IBP and novel GGDPS inhibitors such as RAM2061 have been shown to have key drug-like features including prolonged half-life, metabolic stability, and systemic distribution. Furthermore, RAM2061 exerts anti-neoplastic benefits in mouse models of multiple myeloma and Ewing sarcoma. Therefore, we are interested in determining the potential impact of RAM2061 on osteoclast biology and bone remodeling. Studies utilizing undifferentiated RAW264.7 cells demonstrated that treatment with RAM2061 depletes cells of geranylgeranyl diphosphate, impairs protein geranylgeranylation, and induces markers of the unfolded protein response pathway and apoptosis. Differentiation of RAW264.7 cells to mature osteoclasts is disrupted by RAM2061, resulting in decreased numbers of mature osteoclasts, altered morphology, and decreased tartrate-resistant acid phosphatase activity. Treatment of fully differentiated RAW264.7 cells with RAM2061 led to decreased resorptive activity. Confocal microscopy studies revealed that RAM2061 disrupts Cdc42 localization, inhibiting proper actin ring formation in osteoclasts. No significant impact on bone turnover markers or bone histomorphology was observed following a 3-week treatment of CD-1 mice with RAM2061, although decreased numbers of osteoclasts were observed. Liquid chromatography-tandem mass spectrometry studies confirmed accumulation of RAM2061 in bone from the in vivo studies as well as hydroxyapatite binding in vitro. In conclusion, these studies are the first to demonstrate the anti-osteoclastic activity of GGDPS inhibitor treatment and support future studies exploring the therapeutic benefit of this novel therapy in the setting of pathological bone remodeling.
Keywords: antiresorptives; molecular pathways—remodeling; osteoclasts; other (therapeutics); preclinical studies.
© The Author(s) 2024. Published by Oxford University Press on behalf of the American Society for Bone and Mineral Research.
Conflict of interest statement
The authors have no conflicts of interest.
Figures




Similar articles
-
In Vivo Evaluation of Isoprenoid Triazole Bisphosphonate Inhibitors of Geranylgeranyl Diphosphate Synthase: Impact of Olefin Stereochemistry on Toxicity and Biodistribution.J Pharmacol Exp Ther. 2019 Nov;371(2):327-338. doi: 10.1124/jpet.119.258624. Epub 2019 Aug 16. J Pharmacol Exp Ther. 2019. PMID: 31420526 Free PMC article.
-
Geranylgeranyl diphosphate synthase inhibitor and proteasome inhibitor combination therapy in multiple myeloma.Exp Hematol Oncol. 2022 Feb 9;11(1):5. doi: 10.1186/s40164-022-00261-6. Exp Hematol Oncol. 2022. PMID: 35139925 Free PMC article.
-
Preclinical investigation of a potent geranylgeranyl diphosphate synthase inhibitor.Invest New Drugs. 2018 Oct;36(5):810-818. doi: 10.1007/s10637-018-0571-3. Epub 2018 Mar 2. Invest New Drugs. 2018. PMID: 29497895 Free PMC article.
-
A patent review of bisphosphonates in treating bone disease.Expert Opin Ther Pat. 2019 May;29(5):315-325. doi: 10.1080/13543776.2019.1608180. Epub 2019 Apr 25. Expert Opin Ther Pat. 2019. PMID: 31023104 Review.
-
Recent Advances in the Development of Mammalian Geranylgeranyl Diphosphate Synthase Inhibitors.Molecules. 2017 May 27;22(6):886. doi: 10.3390/molecules22060886. Molecules. 2017. PMID: 28555000 Free PMC article. Review.
Cited by
-
The role of the unfolded protein response pathway in bone homeostasis and potential therapeutic target in cancer-associated bone disease.Bone Res. 2025 Aug 28;13(1):76. doi: 10.1038/s41413-025-00457-6. Bone Res. 2025. PMID: 40877255 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

