Assessment of NanoString technology as a tool for profiling circulating miRNA in maternal blood during pregnancy
- PMID: 39697629
- PMCID: PMC11648433
- DOI: 10.20517/evcna.2024.38
Assessment of NanoString technology as a tool for profiling circulating miRNA in maternal blood during pregnancy
Abstract
Aim: Circulating maternal MicroRNA (miRNA) is a promising source of biomarkers for antenatal diagnostics. NanoString nCounter is a popular global screening tool due to its simplicity and ease of use, but there is a lack of standardisation in analysis methods. We examined the effect of user-defined variables upon reported changes in maternal blood miRNA during pregnancy.
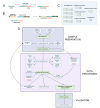
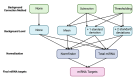
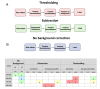
Methods: Total RNA was prepared from the maternal blood of pregnant and control rats. miRNA expression was profiled using Nanostring nCounter. Raw count data were processed using nSolver using different combinations of normalisation and background correction methods as well as various background thresholds. A panel of 14 candidates in which changes were supported by multiple analysis workflows was selected for validation by RT-qPCR. We then reverse-engineered the nSolver analysis to gain further insight.
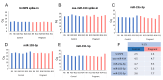
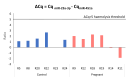
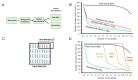
Results: Thirty-one putative differentially expressed miRNAs were identified by nSolver. However, each analysis workflow produced a different set of reported biomarkers and none of them was common to all analysis methods. Four miRNAs with known roles in pregnancy (miR-183, miR-196c, miR-431, miR-450a) were validated. No single nSolver analysis workflow could successfully identify all four validated changes. Reverse engineering revealed errors in nSolver data processing which compound the inherent problems associated with background correction and normalisation.
Conclusion: Our results suggest that user-defined variables greatly influence the output of the assay. This highlights the need for standardised nSolver data analysis methods and detailed reporting of these methods. We suggest that investigators in the future should not rely on a single analysis method to identify changes and should always validate screening results.
Keywords: Micro RNA; blood biomarker; expression profiling; pregnancy.
© The Author(s) 2024.
Conflict of interest statement
All authors declared that there are no conflicts of interest.
Figures



Similar articles
-
Identification of reference miRNAs in plasma useful for the study of oestrogen-responsive miRNAs associated with acquired Protein S deficiency in pregnancy.BMC Res Notes. 2017 Jul 25;10(1):312. doi: 10.1186/s13104-017-2636-3. BMC Res Notes. 2017. PMID: 28743297 Free PMC article.
-
Analysis of circulating microRNA during early gestation in Japanese black cattle.Domest Anim Endocrinol. 2022 Apr;79:106706. doi: 10.1016/j.domaniend.2021.106706. Epub 2021 Dec 5. Domest Anim Endocrinol. 2022. PMID: 34973621
-
G2/M Checkpoint Modulation: Insights from miRNA Profiles in FAM and Breast Cancer.Asian Pac J Cancer Prev. 2024 Aug 1;25(8):2661-2668. doi: 10.31557/APJCP.2024.25.8.2661. Asian Pac J Cancer Prev. 2024. PMID: 39205563 Free PMC article.
-
Challenges and opportunities in processing NanoString nCounter data.Comput Struct Biotechnol J. 2024 Apr 30;23:1951-1958. doi: 10.1016/j.csbj.2024.04.061. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38736697 Free PMC article. Review.
-
MicroRNA Expression Profiling to Identify and Validate Reference Genes for the Relative Quantification of microRNA in Rectal Cancer.PLoS One. 2016 Mar 3;11(3):e0150593. doi: 10.1371/journal.pone.0150593. eCollection 2016. PLoS One. 2016. PMID: 26937645 Free PMC article.
References
-
- Ferentzi H, Pfitzer C, Rosenthal LM, Berger F, Schmitt KRL. Long-term early development research in congenital heart disease (LEADER-CHD): a study protocol for a prospective cohort observational study investigating the development of children after surgical correction for congenital heart defects during the first 3 years of life. BMJ Open. 2017;7:e018966. doi: 10.1136/bmjopen-2017-018966. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
