Identification of a novel mycovirus belonging to the "flexivirus"-related family with icosahedral virion
- PMID: 39697687
- PMCID: PMC11654247
- DOI: 10.1093/ve/veae093
Identification of a novel mycovirus belonging to the "flexivirus"-related family with icosahedral virion
Abstract
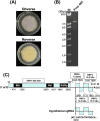
The order Tymovirales currently comprises five viral families with positive-sense RNA [(+)RNA] genomes that infect plants, fungi, and insects. Virion morphologies within the order Tymovirales differ between families, with icosahedral virions in the Tymoviridae and filamentous virions in the other "flexi"viridae families. Despite their different morphologies, these viruses are placed in the same order based on phylogenetic analyses of replicase-associated polyproteins. However, one of the families in the Tymovirales, Deltaflexiviridae, is considered to be capsidless because there have been no published reports of virion isolation. Here, we report that a new "flexivirus"-related (+)RNA virus, prospectively named Fusarium oxysporum icosahedral virus 1 (FoIV1), is icosahedral and that most deltaflexiviruses may have icosahedral virions. Phylogenetic analyses based on replicase-associated polyproteins indicated that FoIV1 forms a distinct group in the Tymovirales with some viruses originally assigned to the Deltaflexiviridae. Electron microscopy, protein analysis, and protein structure predictions indicate that FoIV1 open reading frame 4 encodes a single jelly-roll (SJR)-like coat protein (CP) that constitutes the icosahedral virions. Results of clustering analyses based on amino acid sequences and predicted CP structures suggested that most of the deltaflexiviruses have icosahedral virions composed of SJR-like CPs as in FoIV1, rather than having filamentous virions or capsidless. These results challenge the conventional understanding of viruses in the order Tymovirales, with important implications for revising its taxonomic framework and providing insights into the evolutionary relationships within this diverse and broad host range group of (+)RNA viruses.
Keywords: Tymovirales; coat protein; deltaflexivirus; mycovirus; single jelly-roll; virus particle.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures





Similar articles
-
Identification of a novel mycovirus isolated from Rhizoctonia solani (AG 2-2 IV) provides further information about genome plasticity within the order Tymovirales.Arch Virol. 2017 Feb;162(2):555-559. doi: 10.1007/s00705-016-3085-3. Epub 2016 Oct 12. Arch Virol. 2017. PMID: 27734236
-
Characterization of a novel Sclerotinia sclerotiorum RNA virus as the prototype of a new proposed family within the order Tymovirales.Virus Res. 2016 Jul 2;219:92-99. doi: 10.1016/j.virusres.2015.11.019. Epub 2015 Nov 18. Virus Res. 2016. PMID: 26603216
-
The complete genome sequence of a novel Fusarium graminearum RNA virus in a new proposed family within the order Tymovirales.Arch Virol. 2016 Oct;161(10):2899-903. doi: 10.1007/s00705-016-2961-1. Epub 2016 Jul 4. Arch Virol. 2016. PMID: 27376377
-
Plantago asiatica mosaic virus: An emerging plant virus causing necrosis in lilies and a new model RNA virus for molecular research.Mol Plant Pathol. 2022 Oct;23(10):1401-1414. doi: 10.1111/mpp.13243. Epub 2022 Jul 20. Mol Plant Pathol. 2022. PMID: 35856603 Free PMC article. Review.
-
[The great virus comeback].Biol Aujourdhui. 2013;207(3):153-68. doi: 10.1051/jbio/2013018. Epub 2013 Dec 13. Biol Aujourdhui. 2013. PMID: 24330969 Review. French.
Cited by
-
Characterization of a multi-segmented rod-shaped mycovirus within the order Martellivirales largely accommodating plant viruses.Virus Res. 2025 Jul;357:199591. doi: 10.1016/j.virusres.2025.199591. Epub 2025 May 30. Virus Res. 2025. PMID: 40451551 Free PMC article.
-
Complete genome sequence and characterisation of a novel flexivirus infecting the necrotrophic conifer pathogen Diplodia sapinea.Arch Virol. 2025 May 16;170(6):131. doi: 10.1007/s00705-025-06313-6. Arch Virol. 2025. PMID: 40379837 Free PMC article.
References
-
- Adams MJ, Candresse T, Hammond J et al. Family—Betaflexiviridae. In: King AMQ, Adams MJ, Carstens EB et al. (eds), Virus Taxonomy, 9th edn. San Diego: Elsevier, 2012a, 920–41.
-
- Adams MJ, Kreuze JF, Martelli GP. Order—Tymovirales. In: King AMQ, Adams MJ, Carstens EB et al. (eds), Virus Taxonomy, 9th edn. San Diego: Elsevier, 2012b, 901–03.
LinkOut - more resources
Full Text Sources
Miscellaneous

