A novel immune-related gene prognostic signature combining immune cell infiltration and immune checkpoint for glioblastoma patients
- PMID: 39697704
- PMCID: PMC11651773
- DOI: 10.21037/tcr-24-562
A novel immune-related gene prognostic signature combining immune cell infiltration and immune checkpoint for glioblastoma patients
Abstract
Background: Glioblastoma (GBM) is a highly lethal brain tumor with a complex tumor microenvironment (TME) and poor prognosis. This study aimed to develop and validate a novel immune-related prognostic model for GBM patients to enhance personalized prognosis prediction and develop effective therapeutic strategies.
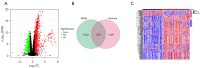
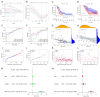
Methods: RNA sequencing and clinical data for GBM patients were obtained from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) (GSE83300). Single-sample gene set enrichment analysis (ssGSEA) was performed using the gene set variation analysis (GSVA) package in R to classify the samples into high and low immune infiltration clusters based on 29 immune cell subtypes. Clustering validations included differential analysis of immune scores and comparison of human leukocyte antigen (HLA) family expression and immune cell subtypes. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Ontology (GO) analysis compared molecular mechanisms and cellular functions between clusters. Differentially expressed immune-related genes between the high and low immune infiltration clusters were screened out, and the prognostic immune-related genes (PIGs) were identified using univariate Cox regression. Co-expression analysis between PIGs and transcription factors (TFs) (Cistrome) was conducted, and a protein-protein interaction (PPI) network (STRING) was constructed. Least absolute shrinkage and selection operator (LASSO) regression constructed a prognostic model. Correlation analyses between PIGs, immune infiltrates, and GBM-related genes were performed. Tumor mutation burden (TMB) analysis and a nomogram incorporating age, gender, and risk score were developed for individualized prognosis prediction.
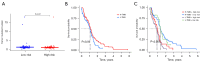
Results: A total of 312 differentially expressed immune-related genes were identified between high and low immune infiltration clusters. Of these, 28 genes were correlated with GBM prognosis. LASSO regression identified 10 genes (CLCF1, PTX3, TNFRSF14, SDC2, VGF, AREG, PLAUR, GRN, AQP9, and IGLV6-57) for the prognostic model. Patients were divided into high-risk and low-risk groups based on risk scores. Survival analysis showed significantly better overall survival (OS) for the low-risk group (P<0.05). The prognostic signature was validated as an independent prognostic factor. Correlation analyses demonstrated significant associations between the prognostic model, immune cell infiltrates, GBM-related genes, and immune checkpoint-related genes. A nomogram incorporating age, gender, and risk score was developed for personalized prognosis prediction.
Conclusions: In summary, our study provided a novel prognostic model based on ssGSEA for GBM patients and offered potential insights for understanding the tumor immune and molecular mechanisms of the disease.
Keywords: Glioblastoma (GBM); gene; immune infiltration; prognostic model; single-sample gene set enrichment analysis (ssGSEA).
2024 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-562/coif). The authors have no conflicts of interest to declare.
Figures






References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
