N-acetyltransferase 10 as a novel prognostic biomarker in papillary renal cell carcinoma: a machine learning and experimental validation study
- PMID: 39697710
- PMCID: PMC11651807
- DOI: 10.21037/tcr-2024-2195
N-acetyltransferase 10 as a novel prognostic biomarker in papillary renal cell carcinoma: a machine learning and experimental validation study
Abstract
Background: N-acetyltransferase 10 (NAT10) is a lysine acetyltransferase known for catalyzing the N4-acetylcytidine (ac4C) modifications on RNAs. Recent studies have associated NAT10 with the pathogenesis of various cancers. However, its specific function and prognostic significance in papillary renal cell carcinoma (pRCC) remain poorly understood. This study aimed to explore NAT10's prognostic value and mechanisms in pRCC.
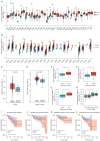
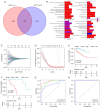
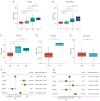
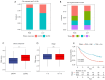
Methods: NAT10 expression and prognostic associations in pancancer were analyzed using The Cancer Genome Atlas (TCGA). Single-cell RNA (scRNA) sequencing data from a previous study were used to characterize NAT10 expression at the single-cell level in pRCC. Pathway enrichment analysis, including gene set variance analysis (GSVA) and overrepresentation analysis, was conducted to investigate the potential mechanisms through which NAT10 exerts its effects. Immune cell infiltration analysis, conducted through the ESTIMATE and CIBERSORT algorithms, was performed to examine the association of NAT10 with the tumor microenvironment (TME). A NAT10-related prognostic model was constructed using least absolute shrinkage and selection operator (LASSO) Cox regression on genes that were both positively correlated with NAT10 and were identified as NAT10-mediated ac4C targets by ac4C RNA immunoprecipitation sequencing The model's performance was validated in the TCGA training set (n=285), with 42 events (deaths) and 243 censored cases, and in the GSE2748 external validation set (n=28), with 13 events (deaths) and 15 censored cases. The association between NAT10-related risk scores and immunotherapy response was assessed via the IMvigor210 cohort. Finally, the aberrant expression of NAT10 was confirmed through immunohistochemistry data from the Human Protein Atlas database and our experimental validations from quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot analyses.
Results: NAT10 is upregulated in multiple cancers, including pRCC, and higher NAT10 expression correlates with advanced stages and poorer prognosis. Single-cell RNA-sequencing data confirmed elevated NAT10 expression in malignant pRCC cells. Pathway enrichment and immune cell infiltration analyses indicated that NAT10 is associated with malignancy-related pathways and a disordered TME. A prognostic model was constructed using LASSO Cox regression, with 18 core genes being identified, and demonstrated high predictive accuracy for survival. The model achieved AUC values of 0.97, 0.93, and 0.82 for 1-, 3-, and 5-year survival in the TCGA training set, respectively, while all AUC values in the GSE2748 external validation set were 1. Higher NAT10-related risk scores were linked to poorer immunotherapy response in the IMvigor210 cohort. NAT10's prognostic significance was validated across various cancers, with elevated expression at both the messenger RNA and protein levels confirmed through immunohistochemistry and experimental validation in RCC cell lines.
Conclusions: Our findings suggest that NAT10 is aberrantly expressed in pRCC, is associated with poor prognosis, and contributes to pRCC progression through multiple pathways, offering new insights into the personalized treatment of patients with pRCC.
Keywords: N-acetyltransferase 10 (NAT10); RNA modification; machine learning; papillary renal cell carcinoma (pRCC).
2024 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2024-2195/coif). All authors report that this work was supported by the Natural Science Foundation of Zhejiang Province (No. LY22H160009). The authors have no other conflicts of interest to declare.
Figures

Similar articles
-
Cuproptosis-Related LncRNA-Based Prediction of the Prognosis and Immunotherapy Response in Papillary Renal Cell Carcinoma.Int J Mol Sci. 2023 Jan 11;24(2):1464. doi: 10.3390/ijms24021464. Int J Mol Sci. 2023. PMID: 36674979 Free PMC article.
-
Targeting N4-acetylcytidine suppresses hepatocellular carcinoma progression by repressing eEF2-mediated HMGB2 mRNA translation.Cancer Commun (Lond). 2024 Sep;44(9):1018-1041. doi: 10.1002/cac2.12595. Epub 2024 Jul 19. Cancer Commun (Lond). 2024. PMID: 39030964 Free PMC article.
-
Integrated Bioinformatics and Experimental Validation to Identify a Disulfidptosis-Related lncRNA Model for Prognostic Prediction in Papillary Renal Cell Carcinoma.Comb Chem High Throughput Screen. 2025;28(5):883-898. doi: 10.2174/0113862073303084240403051346. Comb Chem High Throughput Screen. 2025. PMID: 38639274
-
Emerging role of N-acetyltransferase 10 in diseases: RNA ac4C modification and beyond.Mol Biomed. 2025 Jul 1;6(1):46. doi: 10.1186/s43556-025-00286-3. Mol Biomed. 2025. PMID: 40588654 Free PMC article. Review.
-
Emerging Role of NAT10 as ac4C Writer in Inflammatory Diseases: Mechanisms and Therapeutic Applications.Curr Drug Targets. 2025;26(4):282-294. doi: 10.2174/0113894501346709241202110834. Curr Drug Targets. 2025. PMID: 39633518 Review.
References
LinkOut - more resources
Full Text Sources
