Prognostic signature detects homologous recombination deficient in glioblastoma
- PMID: 39697739
- PMCID: PMC11651736
- DOI: 10.21037/tcr-23-2077
Prognostic signature detects homologous recombination deficient in glioblastoma
Abstract
Background: Glioblastoma (GBM) is a frequent malignant tumor in neurosurgery characterized by a high degree of heterogeneity and genetic instability. DNA double-strand breaks generated by homologous recombination deficiency (HRD) are a well-known contributor to genomic instability, which can encourage tumor development. It is unknown, however, whether the molecular characteristics linked with HRD have a predictive role in GBM. The study aims to assess the extent of genomic instability in GBM using HRD score and investigate the prognostic significance of HRD-related molecular features in GBM.
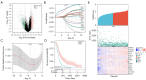
Methods: The discovery cohort comprised 567 GBM patients from The Cancer Genome Atlas (TCGA) database. We established HRD scores using the single nucleotide polymorphism (SNP) array data and analyzed transcriptomic data from patients with different HRD scores to identify biomarkers associated with HRD. A prognostic model was built by using HRD-related differentially expressed genes (DEGs) and validated in a distinct cohort from the Chinese Glioma Genome Atlas (CGGA) database.
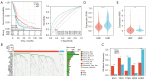
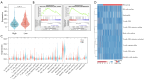
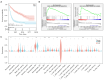
Results: Based on the SNP array data, the gene expression profile data, and the clinical characteristics of GBM patients, we found that patients with a high HRD score had a better prognosis than those with a low HRD score. The DNA damage repair (DDR) signaling pathways were notably enriched in the HRD-positive subgroup. The prognostic model was developed by including HRD-related DEGs that could evaluate the clinical prognosis of patients more efficiently than the HRD score. In addition, patients with a low-risk score had a considerably augmented signature of γδT cells. Finally, through univariate and multivariate Cox regression analyses, it was demonstrated that the prognostic model was superior to other prognostic markers.
Conclusions: In conclusion, our research has not only demonstrated that a high HRD score is a valid prognostic biomarker in GBM patients but also built a stable prognosis model [odds ratio (OR) 0.18, 95% confidence interval (CI): 0.11-0.23, P<0.001] that is more accurate than conventional prognostic markers such as O6-methylguanine-DNA methyltransferase (MGMT) methylation (OR 0.55, 95% CI: 0.33-0.91, P=0.02).
Keywords: Glioblastoma (GBM); homologous recombination deficient (HRD); prognostic factor; risk scoring model.
2024 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2077/coif). The authors have no conflicts of interest to declare.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
