Integrative multi-omics and machine learning approach reveals tumor microenvironment-associated prognostic biomarkers in ovarian cancer
- PMID: 39697754
- PMCID: PMC11651752
- DOI: 10.21037/tcr-24-539
Integrative multi-omics and machine learning approach reveals tumor microenvironment-associated prognostic biomarkers in ovarian cancer
Abstract
Background: Ovarian cancer (OC) is a globally prevalent malignancy with significant morbidity and mortality, yet its heterogeneity poses challenges in treatment and prognosis. Recognizing the crucial role of the tumor microenvironment (TME) in OC progression, this study leverages integrative multi-omics and machine learning to uncover TME-associated prognostic biomarkers, paving the way for more personalized therapeutic interventions.
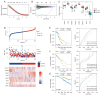
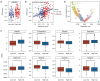
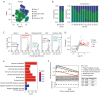
Methods: Employing a rigorous multi-omics approach, this study analyzed single-cell RNA sequencing (scRNA-seq) data from OC and normal tissue samples, including high-grade serous OC (HGSOC) from the Gene Expression Omnibus (GEO: GSE184880) and The Cancer Genome Atlas (TCGA) OC cohort, utilizing the Seurat package to annotate 700 TME-related genes. A prognostic model was developed using the least absolute shrinkage and selection operator (LASSO) regression and independently validated against similarly composed HGSOC datasets. Comprehensive gene expression and immune cell infiltration analyses were conducted, employing advanced algorithms like xCell to delineate the immune landscape of HGSOC.
Results: Our investigation unveiled distinctive immune cell infiltration patterns and gene expression profiles within the TME of HGSOC. Notably, the prevalence of exhausted CD8+ T cells in high-risk patient samples emerged as a critical finding, underscoring the dualistic nature of the immune response in OC. The developed prognostic model, incorporating immune cell markers, exhibited robust predictive accuracy for patient outcomes, showing significant correlations with immunotherapy responses and drug sensitivities.
Conclusions: This study presents a groundbreaking exploration of the OC TME, offering vital insights into its molecular intricacies. By systematically deciphering the TME-associated gene signatures, the research illuminates the potential of these biomarkers in refining patient prognosis and guiding treatment strategies. Our findings underscore the necessity for personalized medicine in OC treatment, potentially enhancing patient survival rates and quality of life. This study marks a significant stride in understanding and combatting the complexities of OC.
Keywords: Ovarian cancer (OC); machine learning; prognostic biomarkers; single-cell RNA sequencing (scRNA-seq); tumor microenvironment (TME).
2024 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-539/coif). The authors have no conflicts of interest to declare.
Figures






References
-
- Mazidimoradi A, Momenimovahed Z, Khani Y, et al. Global patterns and temporal trends in ovarian cancer morbidity, mortality, and burden from 1990 to 2019. Oncologie 2023;25:641-59. 10.1515/oncologie-2023-0172 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
