Unveiling the hidden diversity and functional role of Chloroflexota in full-scale wastewater treatment plants through genome-centric analyses
- PMID: 39698295
- PMCID: PMC11653643
- DOI: 10.1093/ismeco/ycae050
Unveiling the hidden diversity and functional role of Chloroflexota in full-scale wastewater treatment plants through genome-centric analyses
Abstract
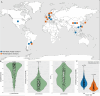
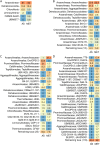
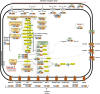
The phylum Chloroflexota has been found to exhibit high abundance in the microbial communities from wastewater treatment plants (WWTPs) in both aerobic and anaerobic systems. However, its metabolic role has not been fully explored due to the lack of cultured isolates. To address this gap, we use publicly available metagenome datasets from both activated sludge (AS) and methanogenic (MET) full-scale wastewater treatment reactors to assembled genomes. Using this strategy, 264 dereplicated, medium- and high-quality metagenome-assembled genomes (MAGs) classified within Chloroflexota were obtained. Taxonomic classification revealed that AS and MET reactors harbored distinct Chloroflexota families. Nonetheless, the majority of the annotated MAGs (166 MAGs with >85% completeness and < 5% contamination) shared most of the metabolic potential features, including the ability to degrade simple sugars and complex polysaccharides, fatty acids and amino acids, as well as perform fermentation of different products. While Chloroflexota MAGs from MET reactors showed the potential for strict fermentation, MAGs from AS harbored the potential for facultatively aerobic metabolism. Metabolic reconstruction of Chloroflexota members from AS unveiled their versatile metabolism and suggested a primary role in hydrolysis, carbon removal and involvement in nitrogen cycling, thus establishing them as fundamental components of the ecosystem. Microbial reference genomes are essential resources for understanding the potential functional role of uncultured organisms in WWTPs. Our study provides a comprehensive genome catalog of Chloroflexota for future analyses aimed at elucidating their role in these ecosystems.
Keywords: Chloroflexota; activated sludge; meta-analysis; metagenome assembled genomes; methanogenic reactors.
© The Author(s) 2024. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Verstraete W, Morgan-Sagastume F, Aiyuk Set al. Anaerobic digestion as a core technology in sustainable management of organic matter. Water Sci Technol 2005;52:59–66. 10.2166/wst.2005.0498Available from. https://iwaponline.com/wst/article-pdf/52/1-2/59/433904/59.pdf - DOI - PubMed
-
- Zamri MFMA, Hasmady S, Akhiar Aet al. A comprehensive review on anaerobic digestion of organic fraction of municipal solid waste. Renew Sust Energ Rev 2021;137:110637. 10.1016/j.rser.2020.110637 - DOI
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

