A comparative analysis of small extracellular vesicle (sEV) micro-RNA (miRNA) isolation and sequencing procedures in blood plasma samples
- PMID: 39698410
- PMCID: PMC11648519
- DOI: 10.20517/evcna.2023.55
A comparative analysis of small extracellular vesicle (sEV) micro-RNA (miRNA) isolation and sequencing procedures in blood plasma samples
Abstract
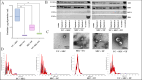
Aims: Analysis of miRNA (18-23nt) encapsulated in small extracellular vesicles (sEVs) (diameter ~30-200 nm) is critical in understanding the diagnostic and therapeutic value of sEV miRNA. However, various sEV enrichment techniques yield different quantities and qualities of sEV miRNA. Here, we compare the efficacy of three sEV isolation techniques in four combinations for miRNA next-generation sequencing.
Methods: Blood plasma from four Holstein-Friesian dairy cows (Bos taurus) (n = 4) with similar genetic traits and physical characteristics were pooled to isolate sEV. Ultracentrifugation (UC) (100,000 × g, 2 h at 4 °C), size-exclusion chromatography (SEC) and ultrafiltration (UF) were used to design four groups of sEV isolations (UC+SEC, SEC+UC, SEC+UF and UC+SEC+UF). sEV miRNAs were isolated using a combination of TRIzol, Chloroform and miRNeasy mini kit (n = 4/each), later sequenced utilizing Novaseq S1 platform (single-end 100 bp sequencing).
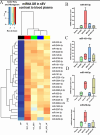
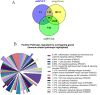
Results: All four sEV methods yielded > 1,700 miRNAs and sEV miRNAs demonstrated a clear separation from control blood plasma circulating miRNA (PCA analysis). MiR-381-3p, miR-23-3p, and miR-18b-3p are among the 25 miRNAs unique to sEV, indicating potential sEV-specific miRNA markers. Further, those 25 miRNAs mostly regulate immune-related functions, indicating the value of sEV miRNA cargo in immunology.
Conclusion: The four sEV miRNA isolation methods employed in this study are valid techniques. The choice of method depends on the research question and study design. If purity is of concern, the UC+SEC method resulted in the best particles/µg protein ratio, which is often used as an indication of sample purity. These results could eventually establish sEV miRNAs as effective diagnostic and therapeutic tools of immunology.
Keywords: Small extracellular vesicles; exosomes; immune function; miRNA; next-generation sequencing; size exclusion chromatography; ultracentrifugation; ultrafiltration.
© The Author(s) 2024.
Conflict of interest statement
All authors declared that there are no conflicts of interest.
Figures





References
LinkOut - more resources
Full Text Sources
