Anti-cancer role of curcumin in prostate cancer cells via regulation of m6A-modified circ0030568-FMR1 signaling pathway
- PMID: 39698574
- PMCID: PMC11650353
- DOI: 10.21037/tau-24-276
Anti-cancer role of curcumin in prostate cancer cells via regulation of m6A-modified circ0030568-FMR1 signaling pathway
Abstract
Background: Prostate cancer (PCa) is the most prevalent adult malignancies worldwide and studies have shown that circular RNAs (circRNAs) play critical roles in the development and progression of PCa. As the most abundant modification, N6-methyladenosine (m6A) modification functions in regulating circRNAs expression and has been shown to regulate PCa progression. However, the biological relevance of m6A modification of circRNAs in PCa remains unclear. In addition, curcumin is reported to inhibit a variety of cancer cells while the biological functions in PCa have not yet been fully elucidated. Thus, our study aims to investigate whether curcumin can suppress PCa progression through the m6A-modified circRNAs.
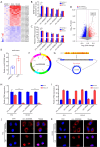
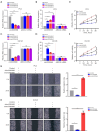
Methods: By conducting m6A methylation immunoprecipitation combined with quantitative real-time polymerase chain reaction (MeRIP-qPCR) assay, cell counting kit-8 (CCK-8) assay and wound healing assay, increased m6A modification on circ0030568 was detected and upregulated circ0030568 was also observed in different PCa cells lines, which promotes proliferation and migration of PCa cells.
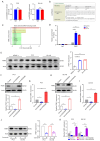
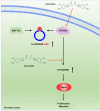
Results: More importantly, the results confirmed that curcumin could suppress the proliferation and migration of PCa cells lines by inhibiting METTL3-modified circ0030568. Mechanistically, m6A reader YTHDF2 elevated the stability of circ0030568 via m6A modification and curcumin could suppress PCa progression by inhibiting YTHDF2 mediated circ0030568 stability.
Conclusions: Taken together, circ0030568 may act as a promising biomarker and an attractive target for PCa treatment and curcumin's inhibition of m6A-modified circ0030568 may be a potential mechanism of its anti-PCa.
Keywords: Curcumin; FMR1; N6-methyladenosine (m6A); circ0030568; prostate cancer (PCa).
2024 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-24-276/coif). The authors have no conflicts of interest to declare.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
