Structural plasticity of the coiled-coil interactions in human SFPQ
- PMID: 39698821
- PMCID: PMC11754644
- DOI: 10.1093/nar/gkae1198
Structural plasticity of the coiled-coil interactions in human SFPQ
Abstract
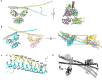
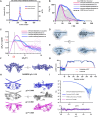
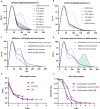
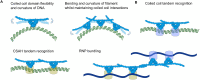
The proteins SFPQ (splicing Factor Proline/Glutamine rich) and NONO (non-POU domain-containing octamer-binding protein) are mammalian members of the Drosophila Behaviour/Human Splicing (DBHS) protein family, which share 76% sequence identity in their conserved 320 amino acid DBHS domain. SFPQ and NONO are involved in all steps of post-transcriptional regulation and are primarily located in mammalian paraspeckles: liquid phase-separated, ribonucleoprotein sub-nuclear bodies templated by NEAT1 long non-coding RNA. A combination of structured and low-complexity regions provide polyvalent interaction interfaces that facilitate homo- and heterodimerisation, polymerisation, interactions with oligonucleotides, mRNA, long non-coding RNA, and liquid phase-separation, all of which have been implicated in cellular homeostasis and neurological diseases including neuroblastoma. The strength and competition of these interaction modes define the ability of DBHS proteins to dissociate from paraspeckles to fulfil functional roles throughout the nucleus or the cytoplasm. In this study, we define and dissect the coiled-coil interactions which promote the polymerisation of DBHS proteins, using a crystal structure of an SFPQ/NONO heterodimer which reveals a flexible coiled-coil interaction interface which differs from previous studies. We support this through extensive solution small-angle X-ray scattering experiments using a panel of SFPQ/NONO heterodimer variants which are capable of tetramerisation to varying extents. The QM mutant displayed a negligible amount of tetramerisation (quadruple loss of function coiled-coil mutant L535A/L539A/L546A/M549A), the Charged Single Alpha Helix (ΔCSAH) variant displayed a dimer-tetramer equilibrium interaction, and the disulfide-forming variant displayed constitutive tetramerisation (R542C which mimics the pathological Drosophila nonAdiss allele). We demonstrate that newly characterised coiled-coil interfaces play a role in the polymerisation of DBHS proteins in addition to the previously described canonical coiled-coil interface. The detail of these interactions provides insight into a process critical for the assembly of paraspeckles as well as the behaviour of SFPQ as a transcription factor, and general multipurpose auxiliary protein with functions essential to mammalian life. Our understanding of the coiled coil behaviour of SFPQ also enhances the explanatory power of mutations (often disease-associated) observed in the DBHS family, potentially allowing for the development of future medical options such as targeted gene therapy.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Structural dynamics of IDR interactions in human SFPQ and implications for liquid-liquid phase separation.Acta Crystallogr D Struct Biol. 2025 Jul 1;81(Pt 7):357-379. doi: 10.1107/S2059798325005303. Epub 2025 Jun 27. Acta Crystallogr D Struct Biol. 2025. PMID: 40574713 Free PMC article.
-
Crystal structure of a SFPQ/PSPC1 heterodimer provides insights into preferential heterodimerization of human DBHS family proteins.J Biol Chem. 2018 Apr 27;293(17):6593-6602. doi: 10.1074/jbc.RA117.001451. Epub 2018 Mar 12. J Biol Chem. 2018. PMID: 29530979 Free PMC article.
-
A new crystal structure and small-angle X-ray scattering analysis of the homodimer of human SFPQ.Acta Crystallogr F Struct Biol Commun. 2019 Jun 1;75(Pt 6):439-449. doi: 10.1107/S2053230X19006599. Epub 2019 May 21. Acta Crystallogr F Struct Biol Commun. 2019. PMID: 31204691 Free PMC article.
-
The Emerging Role of the RNA-Binding Protein SFPQ in Neuronal Function and Neurodegeneration.Int J Mol Sci. 2020 Sep 28;21(19):7151. doi: 10.3390/ijms21197151. Int J Mol Sci. 2020. PMID: 32998269 Free PMC article. Review.
-
Paraspeckles: nuclear bodies built on long noncoding RNA.J Cell Biol. 2009 Sep 7;186(5):637-44. doi: 10.1083/jcb.200906113. Epub 2009 Aug 31. J Cell Biol. 2009. PMID: 19720872 Free PMC article. Review.
Cited by
-
Structural dynamics of IDR interactions in human SFPQ and implications for liquid-liquid phase separation.Acta Crystallogr D Struct Biol. 2025 Jul 1;81(Pt 7):357-379. doi: 10.1107/S2059798325005303. Epub 2025 Jun 27. Acta Crystallogr D Struct Biol. 2025. PMID: 40574713 Free PMC article.
-
Paraspeckle Component 1: a multifunctional RNA binding protein.Am J Cancer Res. 2025 May 25;15(5):2338-2352. doi: 10.62347/RIWH3000. eCollection 2025. Am J Cancer Res. 2025. PMID: 40520856 Free PMC article. Review.
References
-
- Fox A.H., Nakagawa S., Hirose T., Bond C.S.. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci. 2018; 43:124–135. - PubMed
-
- Marshall A.C., Cummins J., Kobelke S., Zhu T., Widagdo J., Anggono V., Hyman A., Fox A.H., Bond C.S., Lee M.. Different low-complexity regions of SFPQ play distinct roles in the formation of biomolecular condensates. J. Mol. Biol. 2023; 435:168364. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

