Allele ages provide limited information about the strength of negative selection
- PMID: 39698825
- PMCID: PMC11912868
- DOI: 10.1093/genetics/iyae211
Allele ages provide limited information about the strength of negative selection
Abstract
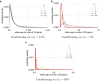
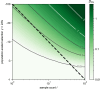
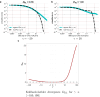
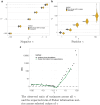
For many problems in population genetics, it is useful to characterize the distribution of fitness effects (DFE) of de novo mutations among a certain class of sites. A DFE is typically estimated by fitting an observed site frequency spectrum (SFS) to an expected SFS given a hypothesized distribution of selection coefficients and demographic history. The development of tools to infer gene trees from haplotype alignments, along with ancient DNA resources, provides us with additional information about the frequency trajectories of segregating mutations. Here, we ask how useful this additional information is for learning about the DFE, using the joint distribution on allele frequency and age to summarize information about the trajectory. To this end, we introduce an accurate and efficient numerical method for computing the density on the age of a segregating variant found at a given sample frequency, given the strength of selection and an arbitrarily complex population size history. We then use this framework to show that the unconditional age distribution of negatively selected alleles is very closely approximated by reweighting the neutral age distribution in terms of the negatively selected SFS, suggesting that allele ages provide little information about the DFE beyond that already contained in the present day frequency. To confirm this prediction, we extended the standard Poisson random field method to incorporate the joint distribution of frequency and age in estimating selection coefficients, and test its performance using simulations. We find that when the full SFS is observed and the true allele ages are known, including ages in the estimation provides only small increases in the accuracy of estimated selection coefficients. However, if only sites with frequencies above a certain threshold are observed, then the true ages can provide substantial information about the selection coefficients, especially when the selection coefficient is large. When ages are estimated from haplotype data using state-of-the-art tools, uncertainty about the age abrogates most of the additional information in the fully observed SFS case, while the neutral prior assumed in these tools when estimating ages induces a downward bias in the case of the thresholded SFS.
Keywords: ARG; DFE; MAF; frequency spectrum; genealogy.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The author(s) declare no conflicts of interest.
Figures

Similar articles
-
Inference of the Distribution of Selection Coefficients for New Nonsynonymous Mutations Using Large Samples.Genetics. 2017 May;206(1):345-361. doi: 10.1534/genetics.116.197145. Epub 2017 Mar 1. Genetics. 2017. PMID: 28249985 Free PMC article.
-
Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies.Genetics. 2007 Dec;177(4):2251-61. doi: 10.1534/genetics.107.080663. Genetics. 2007. PMID: 18073430 Free PMC article.
-
Hill-Robertson interference may bias the inference of fitness effects of new mutations in highly selfing species.Evolution. 2025 Mar 3;79(3):342-363. doi: 10.1093/evolut/qpae168. Evolution. 2025. PMID: 39565285
-
Effects of new mutations on fitness: insights from models and data.Ann N Y Acad Sci. 2014 Jul;1320(1):76-92. doi: 10.1111/nyas.12460. Epub 2014 May 30. Ann N Y Acad Sci. 2014. PMID: 24891070 Free PMC article. Review.
-
A Bayesian method for jointly estimating allele age and selection intensity.Genet Res (Camb). 2008 Feb;90(1):129-37. doi: 10.1017/S0016672307008944. Genet Res (Camb). 2008. PMID: 18289407 Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

